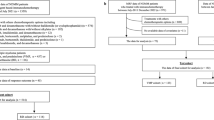
Abstract
This study aimed to establish a predictive model to identify children with hematologic malignancy at high risk for delayed clearance of high-dose methotrexate (HD–MTX) based on machine learning. A total of 205 patients were recruited. Five variables (hematocrit, risk classification, dose, SLC19A1 rs2838958, sex) and three variables (SLC19A1 rs2838958, sex, dose) were statistically significant in univariable analysis and, separately, multivariate logistic regression. The data was randomly split into a “training cohort” and a “validation cohort”. A nomogram for prediction of delayed HD–MTX clearance was constructed using the three variables in the training dataset and validated in the validation dataset. Five machine learning algorithms (cart classification and regression trees, naïve Bayes, support vector machine, random forest, C5.0 decision tree) combined with different resampling methods were used for model building with five or three variables. When developed machine learning models were evaluated in the validation dataset, the C5.0 decision tree combined with the synthetic minority oversampling technique (SMOTE) using five variables had the highest area under the receiver operating characteristic curve (AUC 0.807 [95% CI 0.724–0.889]), a better performance than the nomogram (AUC 0.69 [95% CI 0.594–0.787]). The results support potential clinical application of machine learning for patient risk classification.



Similar content being viewed by others
Data availability
The datasets generated and analyzed during the present study are available from the corresponding author on reasonable request.
Abbreviations
- MTX:
-
Methotrexate
- HD–MTX:
-
High-dose methotrexate
- H:
-
Hour
- SNPs:
-
Single-nucleotide polymorphisms
- ALL:
-
Acute lymphoblastic leukemia
- B-ALL:
-
B cell acute lymphoblastic leukemia
- T-ALL:
-
T cell acute lymphoblastic leukemia
- ANC:
-
Absolute neutrophil count
- PLT:
-
Platelet
- RBC:
-
Red blood cell
- HCT:
-
Hematocrit
- ALT:
-
Alanine aminotransferase
- TBIL:
-
Total bilirubin
- DBIL:
-
Direct bilirubin
- IBIL:
-
Indirect bilirubin
- TP:
-
Total protein
- ALB:
-
Albumin
- GLB:
-
Globulin
- Cr:
-
Creatinine
- HWE:
-
Hardy–Weinberg equilibrium
- RF:
-
Random forest
- SVM:
-
Support vector machine
- CART:
-
Classification and regression trees
- NB:
-
Naïve Bayes classification
- SMOTE:
-
Synthetic minority oversampling technique
- BLSMOTE:
-
Borderline-SMOTE
- ADASYN:
-
Adaptive synthetic
- OSS:
-
One-sided selection
- ROC:
-
Receiver operating characteristic curve
- AUC:
-
Area under the ROC curve
- ACC:
-
Accuracy
- Spec:
-
Specificity
- Sens:
-
Sensitivity
- PPV:
-
Positive predictive value
- NPV:
-
Negative predictive value
- n:
-
Number
References
Stanulla M, Schrappe M. Treatment of childhood acute lymphoblastic leukemia. Semin Hematol. 2009;46:52–63.
Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011;29:551–65.
Levêque D, Santucci R, Gourieux B, Herbrecht R. Pharmacokinetic drug-drug interactions with methotrexate in oncology. Expert Rev Clin Pharmacol. 2011;4:743–50.
Yang SL, Zhao FY, Song H, Shen DY, Xu XJ. Methotrexate associated renal impairment is related to delayed elimination of high-dose methotrexate. Sci World J. 2015;2015:751703.
Ramsey LB, Balis FM, O’Brien MM, Schmiegelow K, Pauley JL, Bleyer A, et al. Consensus guideline for use of glucarpidase in patients with high-dose methotrexate induced acute kidney injury and delayed methotrexate clearance. Oncologist. 2018;23:52–61.
Howard SC, McCormick J, Pui CH, Buddington RK, Harvey RD. Preventing and managing toxicities of high-dose methotrexate. Oncologist. 2016;21:1471–82.
Hu YH, Zhou L, Wang SS, Jing X, Guo HL, Sun F, et al. Methotrexate disposition in pediatric patients with acute lymphoblastic leukemia: what have we learnt from the genetic variants of drug transporters. Curr Pharm Des. 2019;25:627–34.
Cao M, Guo M, Wu DQ, Meng L. Pharmacogenomics of methotrexate: current status and future outlook. Curr Drug Metab. 2018;19:1182–7.
Gervasini G, Mota-Zamorano S. Clinical Implications of methotrexate pharmacogenetics in childhood acute lymphoblastic leukaemia. Curr Drug Metab. 2019;20:313–30.
Ranchon F, Vantard N, Henin E, Bachy E, Sarkozy C, Karlin L, et al. Delayed methotrexate elimination: incidence, interaction with antacid drugs, and clinical consequences? Hematol Oncol. 2018;36:399–406.
Schmidt D, Kristensen K, Schroeder H, Wehner PS, Rosthøj S, Heldrup J, et al. Plasma creatinine as predictor of delayed elimination of high-dose methotrexate in childhood acute lymphoblastic leukemia: A Danish population-based study. Pediatr Blood Cancer. 2019;66:e27637.
Kawase A, Yamamoto T, Egashira S, Iwaki M. Stereoselective inhibition of methotrexate excretion by glucuronides of nonsteroidal anti-inflammatory drugs via multidrug resistance proteins 2 and 4. J Pharmacol Exp Ther. 2016;356:366–74.
Suzuki K, Doki K, Homma M, Tamaki H, Hori S, Ohtani H, et al. Co-administration of proton pump inhibitors delays elimination of plasma methotrexate in high-dose methotrexate therapy. Br J Clin Pharmacol. 2009;67:44–9.
Inose R, Takahashi K, Nanno S, Hino M, Nagayama K. Calcium channel blockers possibly delay the elimination of plasma methotrexate in patients receiving high-dose methotrexate therapy. J Chemother. 2019;31:30–4.
Ramsey LB, Mizuno T, Vinks AA, O’Brien MM. Delayed methotrexate clearance in patients with acute lymphoblastic leukemia concurrently receiving dasatinib. Pediatr Blood Cancer. 2019;66:e27618.
Liu G, Xu Y, Wang X, Zhuang X, Liang H, Xi Y, et al. Developing a machine learning system for identification of severe hand, foot, and mouth disease from electronic medical record data. Sci Rep. 2017;7:16341.
Nunnelee JD. Review of an article: the international warfarin pharmacogenetics consortium (2009) Estimation of the warfarin dose with clinical and pharmacogenetic data. NEJM 360(8): 753–764. J Vasc Nurs. 2009;27:109.
Tang J, Liu R, Zhang YL, Liu MZ, Hu YF, Shao MJ, et al. Application of machine-learning models to predict tacrolimus stable dose in renal transplant recipients. Sci Rep. 2017;7:42192.
Csordas K, Lautner-Csorba O, Semsei AF, Harnos A, Hegyi M, Erdelyi DJ, et al. Associations of novel genetic variations in the folate-related and ARID5B genes with the pharmacokinetics and toxicity of high-dose methotrexate in paediatric acute lymphoblastic leukaemia. Br J Haematol. 2014;166:410–20.
Que LP, Huang K, Fang JP, Xu HG, Zhou DH, Li Y, et al. Reassessment of the risk-stratified gd-2008 all protocol. J Pediatr Hematol Oncol. 2018;40:472–7.
Hospira (2011) Label for methotrexate injection. Lake Forest, il: Hospira; available at: https://www.Accessdata.Fda.Gov/drugsatfda_docs/label/2011/011719s117lbl.pdf. Accessed 17 Jan 2020.
Laboratories B (2016) The R project for statistical computing, https://www.R-project.Org/.Accessed 30 Nov 2017. Accessed 30 Nov 2017.
Bunkhumpornpat C, Sinapiromsaran K. CORE: core-based synthetic minority over-sampling and borderline majority under-sampling technique. Int J Data Min Bioinform. 2015;12:44–58.
Li Y, Li H, Yao H. Analysis and study of diabetes follow-up data using a data-mining-based approach in new urban area of urumqi, xinjiang, china, 2016–2017. Comput Math Methods Med. 2018;2018:7207151.
Sufriyana H, Wu YW, Su EC. Prediction of preeclampsia and intrauterine growth restriction: development of machine learning models on a prospective cohort. JMIR Med Inform. 2020;8:e15411.
Siriseriwan W (2019) Smotefamily. A collection of oversampling techniques for class imbalance problem based on smote, https://cran.r-project.org/package=smotefamily. Accessed 21 June 2019.
Pozzolo AD, Olivier C, Bontempi G (2015) Unbalanced: racing for unbalanced methods selection https://cran.r-project.org/package=unbalanced. Accessed 21 June 2019.
Kataoka T, Sakurashita H, Kajikawa K, Saeki Y, Taogoshi T, Matsuo H. Low serum albumin level is a risk factor for delayed methotrexate elimination in high-dose methotrexate treatment. Ann Pharmacother. 2021. https://doi.org/10.1177/1060028021992767.
Nakano T, Kobayashi R, Matsushima S, Hori D, Yanagi M, Suzuki D, et al. Risk factors for delayed elimination of high-dose methotrexate in childhood acute lymphoblastic leukemia and lymphoma. Int J Hematol. 2021;113:744–50.
Skärby T, Jönsson P, Hjorth L, Behrentz M, Björk O, Forestier E, et al. High-dose methotrexate: on the relationship of methotrexate elimination time vs renal function and serum methotrexate levels in 1164 courses in 264 Swedish children with acute lymphoblastic leukaemia (ALL). Cancer Chemother Pharmacol. 2003;51:311–20.
Xu W, Tang Y, Song H, Shi S, Yang S. Retrospective study on elimination delay of methotrexate in high-dose therapy of childhood acute lymphoblastic leukemia in China. J Pediatr Hematol Oncol. 2007;29:688–93.
Xu C, Jackson SA. Machine learning and complex biological data. Genome Biol. 2019;20:76.
Altman N, Krzywinski M. The curse(s) of dimensionality. Nat Methods. 2018;15:399–400.
Kang J, Schwartz R, Flickinger J, Beriwal S. Machine learning approaches for predicting radiation therapy outcomes: a clinician’s perspective. Int J Radiat Oncol Biol Phys. 2015;93:1127–35.
Lundberg SM, Nair B, Vavilala MS, Horibe M, Eisses MJ, Adams T, et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat Biomed Eng. 2018;2:749–60.
Song L, Langfelder P, Horvath S. Random generalized linear model: a highly accurate and interpretable ensemble predictor. BMC Bioinformatics. 2013;14:5.
Marchese Robinson RL, Palczewska A, Palczewski J, Kidley N. Comparison of the predictive performance and interpretability of random forest and linear models on benchmark data sets. J Chem Inf Model. 2017;57:1773–92.
Gregers J, Christensen IJ, Dalhoff K, Lausen B, Schroeder H, Rosthoej S, et al. The association of reduced folate carrier 80G>A polymorphism to outcome in childhood acute lymphoblastic leukemia interacts with chromosome 21 copy number. Blood. 2010;115:4671–7.
Luteijn RD, Zaver SA, Gowen BG, Wyman SK, Garelis NE, Onia L, et al. SLC19A1 transports immunoreactive cyclic dinucleotides. Nature. 2019;573:434–8.
Laverdière C, Chiasson S, Costea I, Moghrabi A, Krajinovic M. Polymorphism G80A in the reduced folate carrier gene and its relationship to methotrexate plasma levels and outcome of childhood acute lymphoblastic leukemia. Blood. 2002;100:3832–4.
Wang SM, Sun LL, Zeng WX, Wu WS, Zhang GL. Effects of a microRNA binding site polymorphism in SLC19A1 on methotrexate concentrations in Chinese children with acute lymphoblastic leukemia. Med Oncol. 2014;31:62.
Lopez-Lopez E, Ballesteros J, Piñan MA, de Toledo JS, Garcia de Andoin N, Garcia-Miguel P, et al. Polymorphisms in the methotrexate transport pathway: a new tool for MTX plasma level prediction in pediatric acute lymphoblastic leukemia. Pharmacogenet Genomics. 2013;23:53–61.
Radtke S, Zolk O, Renner B, Paulides M, Zimmermann M, Möricke A, et al. Germline genetic variations in methotrexate candidate genes are associated with pharmacokinetics, toxicity, and outcome in childhood acute lymphoblastic leukemia. Blood. 2013;121:5145–53.
Liu SG, Gao C, Zhang RD, Zhao XX, Cui L, Li WJ, et al. Polymorphisms in methotrexate transporters and their relationship to plasma methotrexate levels, toxicity of high-dose methotrexate, and outcome of pediatric acute lymphoblastic leukemia. Oncotarget. 2017;8:37761–72.
Kotnik BF, Jazbec J, Grabar PB, Rodriguez-Antona C, Dolzan V. Association between SLC19A1 gene polymorphism and high dose methotrexate toxicity in childhood acute lymphoblastic leukaemia and non Hodgkin malignant lymphoma: introducing a haplotype based approach. Radiol Oncol. 2017;51:455–62.
Xu WQ, Zhang LY, Chen XY, Pan BH, Mao JQ, Song H, et al. Serum creatinine and creatinine clearance for predicting plasma methotrexate concentrations after high-dose methotrexate chemotherapy for the treatment for childhood lymphoblastic malignancies. Cancer Chemother Pharmacol. 2014;73:79–86.
Nader A, Zahran N, Alshammaa A, Altaweel H, Kassem N, Wilby KJ. Population pharmacokinetics of intravenous methotrexate in patients with hematological malignancies: utilization of routine clinical monitoring parameters. Eur J Drug Metab Pharmacokinet. 2017;42:221–8.
Hurkmans EGE, Klumpers MJ, Vermeulen SH, Hagleitner MM, Flucke U, Schreuder HWB, et al. Analysis of drug metabolizing gene panel in osteosarcoma patients identifies association between variants in SULT1E1, CYP2B6 and CYP4F8 and methotrexate levels and toxicities. Front Pharmacol. 2020;11:1241.
Minematsu T, Sugiyama E, Kusama M, Hori S, Yamada Y, Ohtani H, et al. Effect of hematocrit on pharmacokinetics of tacrolimus in adult living donor liver transplant recipients. Transplant Proc. 2004;36:1506–11.
Tharwat A, Moemen YS, Hassanien AE. Classification of toxicity effects of biotransformed hepatic drugs using whale optimized support vector machines. J Biomed Inform. 2017;68:132–49.
Acknowledgements
This work was supported by Grants from the National Natural Scientific Foundation of China (No. 81503166) and the Natural Scientific Foundation of Hunan province in China (2018JJ3846).
Author information
Authors and Affiliations
Contributions
MZ: reviewed the medical records of the patients, participated in the statistical analysis, evaluated the results, and drafted the manuscript. ZC: contributed to the study design, processed the data, and revised the manuscript. CD: conducted machine learning model building. QQ: revised the manuscript. GW: processed the data. SL: reviewed the medical reports of the patients. FW: designed and organized the study, and evaluated the results. All authors discussed and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare no potential conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12185_2021_3184_MOESM2_ESM.tiff
Supplementary Fig. 1. The performance of models developed by machine learning algorithms without resampling in the training dataset: ROC (a); Sensitivity (b); Specificity (c); Accuracy (d). Data presented as box plot showing min, first quartile, median, third quartile, and maximum (TIFF 549 KB)
12185_2021_3184_MOESM3_ESM.tiff
Supplementary Fig. 2. The performance of models developed by machine learning algorithms combination with SMOTE in the training dataset: ROC (a); Sensitivity (b); Specificity (c); Accuracy (d). Data presented as box plot showing min, first quartile, median, third quartile, and maximum (TIFF 552 KB)
12185_2021_3184_MOESM4_ESM.tiff
Supplementary Fig. 3. The performance of predicted models developed by machine learning algorithms combination with ADASYN in the training dataset: ROC (a); Sensitivity (b); Specificity (c); Accuracy (d). Data presented as box plot showing min, first quartile, median, third quartile, and maximum (TIFF 557 KB)
12185_2021_3184_MOESM5_ESM.tiff
Supplementary Fig. 4. The performance of predicted models developed by machine learning algorithms combination with BLSMOTE in the training dataset: ROC (a); Sensitivity (b); Specificity (c); Accuracy (d). Data presented as box plot showing min, first quartile, median, third quartile, and maximum (TIFF 549 KB)
12185_2021_3184_MOESM6_ESM.tiff
Supplementary Fig. 5. The performance of predicted models developed by machine learning algorithms combination with OSS in the training dataset: ROC (a); Sensitivity (b); Specificity (c); Accuracy (d). Data presented as box plot showing min, first quartile, median, third quartile, and maximum (TIFF 548 KB)
About this article
Cite this article
Zhan, M., Chen, Z., Ding, C. et al. Risk prediction for delayed clearance of high-dose methotrexate in pediatric hematological malignancies by machine learning. Int J Hematol 114, 483–493 (2021). https://doi.org/10.1007/s12185-021-03184-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12185-021-03184-w