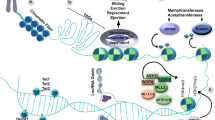
Abstract
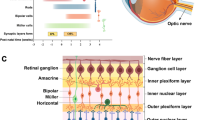
Epigenetic regulation, including DNA methylation, histone modifications, and chromosomal organization, is emerging as a new layer of transcriptional regulation in retinal development and maintenance. Guided by intrinsic transcription factors and extrinsic signaling molecules, epigenetic regulation can activate and/or repress the expression of specific sets of genes, therefore playing an important role in retinal cell fate specification and terminal differentiation during development as well as maintaining cell function and survival in adults. Here, we review the major findings that have linked these mechanisms to the development and maintenance of retinal structure and function, with a focus on ganglion cells and photoreceptors. The mechanisms of epigenetic regulation are highly complex and vary among different cell types. Understanding the basic principles of these mechanisms and their regulatory pathways may provide new insight into the pathogenesis of retinal diseases associated with transcription dysregulation, and new therapeutic strategies for treatment.



Similar content being viewed by others
Abbreviations
- HKM:
-
Histone lysine methylation
- HKMTase:
-
Histone methyltransferase
- HKDMase:
-
Histone lysine demethylase
- DNMTase:
-
DNA methyltransferase
- HAT (KAT):
-
Histone lysine acetyltransferase
- HDAC:
-
Histone deacetylase
- CT:
-
Chromatin territory
- TxF:
-
Transcription factory
References
Humphries MM, et al. Retinopathy induced in mice by targeted disruption of the rhodopsin gene [see comments]. Nat Genet. 1997;15:216–9.
Olsson JE, et al. Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron. 1992;9:815–30.
Guo Y, et al. Early gene expression changes in the retinal ganglion cell layer of a rat glaucoma model. Investig Ophthalmol Vis Sci. 2011;52:1460–73.
Wang DY, et al. Global gene expression changes in rat retinal ganglion cells in experimental glaucoma. Investig Ophthalmol Vis Sci. 2010;51:4084–95.
Panagis L, et al. Gene expression changes in areas of focal loss of retinal ganglion cells in the retina of DBA/2J mice. Investig Ophthalmol Vis Sci. 2010;51:2024–34.
Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–33.
Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–76.
Ohsawa R, Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain research. 2008;1192:90–8.
Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18.
Walton EL, Francastel C, & Velasco G (2011) Maintenance of DNA methylation: Dnmt3b joins the dance. Epigenetics 6.
Peng GH, Chen S. Active opsin loci adopt intrachromosomal loops that depend on the photoreceptor transcription factor network. Proc Natl Acad Sci U S A. 2011;108:17821–6.
Wang Y, et al. A locus control region adjacent to the human red and green visual pigment genes. Neuron. 1992;9:429–40.
Wang Y, et al. Mutually exclusive expression of human red and green visual pigment-reporter transgenes occurs at high frequency in murine cone photoreceptors. Proc Natl Acad Sci U S A. 1999;96:5251–6.
Chakalova L, et al. Developmental regulation of the beta-globin gene locus. Prog Mol Subcell Biol. 2005;38:183–206.
Huang J, et al. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J Virol. 2006;80:5740–6.
Rao RC, et al. Dynamic patterns of histone lysine methylation in the developing retina. Invest Ophthalmol Vis Sci. 2010;51:6784–92.
Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53.
Skowronska-Krawczyk D, Ballivet M, Dynlacht BD, Matter JM. Highly specific interactions between bHLH transcription factors and chromatin during retina development. Development. 2004;131:4447–54.
Tachibana M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91.
Cherrier T, et al. p21(WAF1) gene promoter is epigenetically silenced by CTIP2 and SUV39H1. Oncogene. 2009;28:3380–9.
Eom GH, et al. Histone methyltransferase PRDM8 regulates mouse testis steroidogenesis. Biochem Biophys Res Commun. 2009;388:131–6.
Komai T, Iwanari H, Mochizuki Y, Hamakubo T, Shinkai Y. Expression of the mouse PR domain protein Prdm8 in the developing central nervous system. Gene Expr Patterns. 2009;9:503–14.
Kubicek S, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–81.
Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol. 2005;1:143–5.
Toth Z, et al. Epigenetic analysis of KSHV latent and lytic genomes. PLoS pathogens. 2010;6:e1001013.
Tan J, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–63.
Schermelleh L, et al. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35:4301–12.
Nasonkin IO, et al. Distinct nuclear localization patterns of DNA methyltransferases in developing and mature mammalian retina. J Comp Neurol. 2011;519:1914–30.
Nasonkin IO, et al. (2010) Role of Epigenetics (DNA Methylation) in RPE and Photoreceptor Development. in Invest Ophthalmol Vis Sci (Annual Meeting Abstracts), pp E-Abstract 4304.
Patel K, et al. (2011) The role of Dnmt1 in retinal differentiation. in Invest Ophthalmol Vis Sci (Annual Meeting Abstracts), pp E-Abstract 5982-A5131.
Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57.
Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5:9–15.
Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–7.
McManus KJ, Hendzel MJ. Quantitative analysis of CBP- and P300-induced histone acetylations in vivo using native chromatin. Mol Cell Biol. 2003;23:7611–27.
Jin Q, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–62.
Gaub P, et al. The histone acetyltransferase p300 promotes intrinsic axonal regeneration. Brain. 2011;134:2134–48.
Li X, Kazgan N. Mammalian sirtuins and energy metabolism. Int J Biol Sci. 2011;7:575–87.
Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–93.
Geranton SM (2011) Targeting epigenetic mechanisms for pain relief. Curr Opin Pharmacol.
Pelzel HR, Schlamp CL, Nickells RW. Histone H4 deacetylation plays a critical role in early gene silencing during neuronal apoptosis. BMC Neurosci. 2010;11:62.
Chen B, Cepko CL. Requirement of histone deacetylase activity for the expression of critical photoreceptor genes. BMC Dev Biol. 2007;7:78.
Jensen DE, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–112.
Scheuermann JC, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–7.
Harbour JW, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–3.
Goldstein AM. Germline BAP1 mutations and tumor susceptibility. Nat Genet. 2011;43:925–6.
Lang G, et al. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol Cell Biol. 2011;31:3734–44.
Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–96.
Guillemette B, et al. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS biology. 2005;3:e384.
Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–8.
Cremer T, Cremer M. Chromosome territories. Cold Spring Harbor perspect in biol. 2010;2:a003889.
Hakim O, Sung MH, Hager GL. 3D shortcuts to gene regulation. Curr Opin Cell Biol. 2010;22:305–13.
Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural development. 2011;6:32.
Rapicavoli NA, Poth EM, Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol. 2010;10:49.
Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6.
Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010;7:582–5.
Rapicavoli NA, Blackshaw S. New meaning in the message: noncoding RNAs and their role in retinal development. Dev Dyn. 2009;238:2103–14.
Orom UA, Shiekhattar R. Noncoding RNAs and enhancers: complications of a long-distance relationship. Trends Genet. 2011;27:433–9.
Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6.
Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–94.
Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60.
Chen DF, Schneider GE, Martinou JC, Tonegawa S. Bcl-2 promotes regeneration of severed axons in mammalian CNS. Nature. 1997;385:434–9.
Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–508.
Matter-Sadzinski L, Matter JM, Ong MT, Hernandez J, Ballivet M. Specification of neurotransmitter receptor identity in developing retina: the chick ATH5 promoter integrates the positive and negative effects of several bHLH proteins. Development. 2001;128:217–31.
Solovei I, et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–68.
Sher F, et al. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells. 2008;26:2875–83.
Hirabayashi Y, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–13.
Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–6.
Ding N, et al. MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression. J Biol Chem. 2009;284:2648–56.
Huang S, Shao G, Liu L. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J Biol Chem. 1998;273:15933–9.
Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–13.
Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70.
Laurent L, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–31.
Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep. 2011;12:647–56.
Nguyen CT, et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2'-deoxycytidine. Cancer Res. 2002;62:6456–61.
Xin Z, et al. Role of histone methyltransferase G9a in CpG methylation of the Prader–Willi syndrome imprinting center. J Biol Chem. 2003;278:14996–5000.
Lehnertz B, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–200.
Wang J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–9.
Li J, et al. Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9. PLoS One. 2011;6:e25104.
Chang Y, et al. MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nat Commun. 2011;2:533.
Petkova TD, Seigel GM, Otteson DC. A role for DNA methylation in regulation of EphA5 receptor expression in the mouse retina. Vision Res. 2011;51:260–8.
Bevins N, Lemke G, Reber M. Genetic dissection of EphA receptor signaling dynamics during retinotopic mapping. J Neurosci. 2011;31:10302–10.
Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–52.
Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705.
Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19.
Jones PL, Wolffe AP. Relationships between chromatin organization and DNA methylation in determining gene expression. Semin Cancer Biol. 1999;9:339–47.
Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–92.
Cvekl A, Mitton KP. Epigenetic regulatory mechanisms in vertebrate eye development and disease. Heredity. 2010;105:135–51.
Yanagi Y, Masuhiro Y, Mori M, Yanagisawa J, Kato S. p300/CBP acts as a coactivator of the cone-rod homeobox transcription factor. Biochem Biophys Res Commun. 2000;269:410–4.
Peng GH, Chen S. Crx activates opsin transcription by recruiting HAT-containing co-activators and promoting histone acetylation. Hum Mol Genet. 2007;16:3433–52.
Chen S, et al. Interference of Crx-dependent transcription by ataxin-7 involves interaction between the glutamine regions and requires the ataxin-7 carboxy-terminal region for nuclear localization. Hum Mol Genet. 2004;13:53–67.
Palhan VB, et al. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci U S A. 2005;102:8472–7.
Helmlinger D, et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 2006;4:e67.
Anamika K, et al. Lessons from genome-wide studies: an integrated definition of the coactivator function of histone acetyl transferases. Epigenetics & chromatin. 2010;3:18.
Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–57.
Biermann J, et al. Valproic acid-mediated neuroprotection and regeneration in injured retinal ganglion cells. Invest Ophthalmol Vis Sci. 2010;51:526–34.
Zhang Z, et al. Valproate protects the retina from endoplasmic reticulum stress-induced apoptosis after ischemia-reperfusion injury. Neurosci Lett. 2011;504:88–92.
Biermann J, Boyle J, Pielen A, Lagreze WA. Histone deacetylase inhibitors sodium butyrate and valproic acid delay spontaneous cell death in purified rat retinal ganglion cells. Mol Vis. 2011;17:395–403.
Gaub P, et al. HDAC inhibition promotes neuronal outgrowth and counteracts growth cone collapse through CBP/p300 and P/CAF-dependent p53 acetylation. Cell Death Differ. 2010;17:1392–408.
Clemson CM, et al. Therapeutic potential of valproic acid for retinitis pigmentosa. Br J Ophthalmol. 2011;95:89–93.
Bott M, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–72.
Wiesner T, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–21.
Testa JR, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–5.
Abdel-Rahman MH, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J of Medic Genetics. 2011;48:856–9.
Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8.
Jordan G, Deeb SS, Bosten JM, Mollon JD. The dimensionality of color vision in carriers of anomalous trichromacy. J Vis. 2010;10:12.
Jordan G, Mollon JD. A study of women heterozygous for colour deficiencies. Vis Res. 1993;33:1495–508.
Boatright JH, Nickerson JM, Borst DE. Site-specific DNA hypomethylation permits expression of the IRBP gene. Brain research. 2000;887:211–21.
Bazhin AV, De Smet C, Golovastova MO, Schmidt J, Philippov PP. Aberrant demethylation of the recoverin gene is involved in the aberrant expression of recoverin in cancer cells. Exp Dermatol. 2010;19:1023–5.
Carroll J, et al. Deletion of the X-linked opsin gene array locus control region (LCR) results in disruption of the cone mosaic. Vision Res. 2010;50:1989–99.
Kizilyaprak C, Spehner D, Devys D, Schultz P. The linker histone H1C contributes to the SCA7 nuclear phenotype. Nucleus. 2011;2:444–54.
Peng G-H, Hennig AK, Chen Y, & Chen S (2010) Histone acetyltransferases CBP and p300 in photoreceptors: loss-of-function affects chromatin configuration and transcription of photoreceptor genes ARVO Meeting Abstract 51:1086.
Hennig AK, Wang H, & Chen S (2010) Histone acetyltransferases cbp and p300 are required in rod photoreceptors to maintain retina structure and function ARVO Meeting Abstracts:1085.
Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–31.
Zhao Y, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101.
Zhang XY, et al. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–11.
Abou-Sleymane G, et al. Polyglutamine expansion causes neurodegeneration by altering the neuronal differentiation program. Hum Mol Genet. 2006;15:691–703.
Chen YC, et al. (2011) Gcn5 loss-of-function accelerates cerebellar and retinal degeneration in a SCA7 mouse model. Human molecular genetics.
Bramall AN, Wright AF, Jacobson SG, McInnes RR. The genomic, biochemical, and cellular responses of the retina in inherited photoreceptor degenerations and prospects for the treatment of these disorders. Annu Rev Neurosci. 2010;33:441–72.
Sancho-Pelluz J, et al. Excessive HDAC activation is critical for neurodegeneration in the rd1 mouse. Cell death & disease. 2010;1:e24.
Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009;323:256–9.
Gasser SM, Laemmli UK. The organisation of chromatin loops: characterization of a scaffold attachment site. EMBO J. 1986;5:511–8.
Gasser SM, Laemmli UK. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986;46:521–30.
Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–28.
Marsden MP, Laemmli UK. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979;17:849–58.
Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11.
Wurtele H, Chartrand P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended Chromosome Conformation Capture methodology. Chromosome Res. 2006;14:477–95.
Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7.
Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet. 2006;38:1348–54.
Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–309.
Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93.
Cremer T, et al. Chromosome territoriesa functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–16.
Crutchley JL, Wang XQ, Ferraiuolo MA, Dostie J. Chromatin conformation signatures: ideal human disease biomarkers? Biomarkers in medicine. 2010;4:611–29.
Fraser J, et al. Chromatin conformation signatures of cellular differentiation. Genome Biol. 2009;10:R37.
Cremer M, et al. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods in molecular biol. 2008;463:205–39.
Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61.
Zinner R, Teller K, Versteeg R, Cremer T, Cremer M. Biochemistry meets nuclear architecture: multicolor immuno-FISH for co-localization analysis of chromosome segments and differentially expressed gene loci with various histone methylations. Adv Enzym Regul. 2007;47:223–41.
Boyle AP, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21:456–64.
Asp P, et al. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci U S A. 2011;108:E149–158.
Guo C, et al. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell. 2011;147:332–43.
Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–17.
Noordermeer D, et al. The dynamic architecture of Hox gene clusters. Science. 2011;334:222–5.
He S, et al. Chromatin organization and nuclear microenvironments in cancer cells. J Cell Biochem. 2008;104:2004–15.
Nie Z, Chen S, Kumar R, Zack DJ. RER, an evolutionarily conserved sequence upstream of the rhodopsin gene, has enhancer activity. J Biol Chem. 1996;271:2667–75.
Smallwood PM, Wang Y, Nathans J. Role of a locus control region in the mutually exclusive expression of human red and green cone pigment genes. Proc Natl Acad Sci U S A. 2002;99:1008–11.
de Melo J, Peng GH, Chen S, Blackshaw S. The Spalt family transcription factor Sall3 regulates the development of cone photoreceptors and retinal horizontal interneurons. Development. 2011;138:2325–36.
Hagege H, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc. 2007;2:1722–33.
Kizilyaprak C, Spehner D, Devys D, Schultz P. In vivo chromatin organization of mouse rod photoreceptors correlates with histone modifications. PLoS One. 2010;5:e11039.
Gonzalo S, Blasco MA. Role of Rb family in the epigenetic definition of chromatin. Cell Cycle. 2005;4:752–5.
Eskiw CH, et al. Transcription factories and nuclear organization of the genome. Cold Spring Harbor symposia on quantitative biol. 2010;75:501–6.
Eskiw C, Fraser P. Inverted rod nuclei see the light. Nature cell biol. 2009;11:680–1.
Donovan SL, Schweers B, Martins R, Johnson D, Dyer MA. Compensation by tumor suppressor genes during retinal development in mice and humans. BMC Biol. 2006;4:14.
Macaluso M, et al. Nuclear and cytoplasmic interaction of pRb2/p130 and ER-beta in MCF-7 breast cancer cells. Annals of oncol: Official J of the European Soc for Medic Oncol/ESMO 17. 2006;Suppl 7:vii27–29.
Longworth MS, Dyson NJ. pRb, a local chromatin organizer with global possibilities. Chromosoma. 2010;119:1–11.
Acknowledgments
Our work presented herein was supported by grants from the National Institutes of Health (R01EY017641 to DFC, R01EY012543 to SC, R21DA024803 to DFC, P30EY02687 to WU-DOVS), Department of Veterans Affairs (1I01RX000110 to DFC), and Department of Defense (W81XWH-09-2-0091; W23RYX-9104-N603 to DFC). Additional support was provided by the American Health Foundation (to RCR), a Lew Wasserman Merit Award (to SC), and unrestricted funds from Research to Prevent Blindness (to WU-DOVS).
Author information
Authors and Affiliations
Corresponding author
Additional information
Rajesh C. Rao and Anne K. Hennig contributed equally to this work.
Rights and permissions
About this article
Cite this article
Rao, R.C., Hennig, A.K., Malik, M.T.A. et al. Epigenetic regulation of retinal development and disease. j ocul biol dis inform 4, 121–136 (2011). https://doi.org/10.1007/s12177-012-9083-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12177-012-9083-0