Abstract
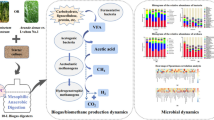
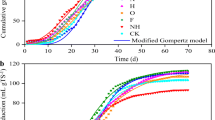
Thermophilic anaerobic digestion (TAD) is an efficient method for biogas production. In this study, the TAD of Arundo donax cv. Lvzhou No. 1 (ADL-1, a new kind of energy crop with high cold tolerance) at different growth stages was carried out, in order to investigate the relationship between microbial community structure and its function during the fermentation process. The results showed that the most optimal growth period of ADL-1 was 3 months, regarding the yield of biogas production. The TAD process lasted for 10 days with cumulative biogas and methane yields of 312.7 mL/g VS and 231 mL/g VS, respectively. The degradation rates of hemicellulose, cellulose, and lignin were 41.78%, 27.99%, and 14.46%, respectively. The high-throughput sequencing of 16S rRNA gene amplicons revealed that the most abundant bacterial phylum in TAD was Firmicutes with three dominant genera of Tepidiphilus, Sedimentibacter, and Gelria. Also, the main archaeal order was Methanomicrobiales, in which Methanoculleus and Methanosarcina were detected as dominant genera. Therefore, this article reveals the dynamic changes of structure and function of microbial communities during TAD of ADL-1, providing the theoretical basis for the development of energy crops with cold tolerance as potential biogas feedstock.






Similar content being viewed by others
References
Deviram G, Mathimani T, Anto S, Ahamed TS, Ananth DA, Pugazhendhi A (2019) Applications of microalgal and cyanobacterial biomass on a way to safe, cleaner and a sustainable environment. J Clean Prod 235:119770. https://doi.org/10.1016/j.jclepro.2019.119770
Saravanan AP, Mathimani T, Deviram G, Rajendran K, Pugazhendhi A (2018) Biofuel policy in India: a review of policy barriers in sustainable marketing of biofuel. J Clean Prod 193. https://doi.org/10.1016/j.jclepro.2018.05.033
Dharmaraja J, Shobana S, Arvindnarayan S, Vadivel M, Atabani AE, Pugazhendhi A, Kumar G (2020) Biobutanol from lignocellulosic biomass: bioprocess strategies. Lignocellulosic Biomass to Liquid Biofuels:169–193. https://doi.org/10.1016/B978-0-12-815936-1.00005-8
Prabakar D, Manimudi VT, Suvetha KS, Sampath S, Mahapatra DM, Rajendran K, Pugazhendhi A (2018) Advanced biohydrogen production using pretreated industrial waste: outlook and prospects. Renew Sust Energ Rev 96:306–324. https://doi.org/10.1016/j.rser.2018.08.006
Anto S, Mukherjee SS, Muthappa R, Mathimani T, Deviram G, Kumar SS, Verma TN, Pugazhendhi A (2019) Algae as green energy reserve: technological outlook on biofuel production. Chemosphere 125079. https://doi.org/10.1016/j.chemosphere.2019.125079
Pugazhendhi A, Mathimani T, Varjani S, Rene ER, Kumar G, Kim SH, Ponnusamy VK, Yoon JJ (2019) Biobutanol as a promising liquid fuel for the future - recent updates and perspectives. Fuel 253:637–646. https://doi.org/10.1016/j.fuel.2019.04.139
Herrmann A (2012) Biogas production from maize: current state, challenges, and prospects. 1. Methane yield potential. Bioenerg Res 5:1027–1042. https://doi.org/10.1007/s12155-012-9202-6
Nges IA, Escobar F, Fu X, Björnsson L (2012) Benefits of supplementing an industrial waste anaerobic digester with energy crops for increased biogas production. Waste Manag 32:0–59. https://doi.org/10.1016/j.wasman.2011.09.009 53
Herrmann C, Idler C, Heiermann M (2016) Biogas crops grown in energy crop rotations: linking chemical composition and methane production characteristics. Bioresour Technol 206:23–35. https://doi.org/10.1016/j.biortech.2016.01.058
Nguyen NS, Soda S, Ishigaki T, Ike M (2012) Microorganisms in landfill bioreactors for accelerated stabilization of solid wastes. J Biosci Bioeng 114:243–250. https://doi.org/10.1016/j.jbiosc.2012.04.007
Li X, Liu Y, Zhang X, Ge C, Piao R, Wang W, Cui Z, Zhao H (2017) Evaluation of biogas production performance and dynamics of the microbial community in different straws. J Microbiol Biotechnol 27:524–534. https://doi.org/10.4014/jmb.1608.08062
Sun W, Sun XX, Cupples AM (2014) Identification of Desulfosporosinus as toluene-assimilating microorganisms from a methanogenic consortium. Int Biodeterior Biodegradation 88:13–19. https://doi.org/10.1016/j.ibiod.2013.11.014
Weiland P (2010) Biogas production: current state and perspectives. Appl Microbiol Biotechnol 85:849–860. https://doi.org/10.1021/ac071042
Azman S, Khadem AF, Lier JBV, Zeeman G, Plugge CM (2015) Presence and role of anaerobic hydrolytic microbes in conversion of lignocellulosic biomass for biogas production. Crit Rev Env Sci Tec 45:2523–2564. https://doi.org/10.1080/10643389.2015.1053727
Hameed SA, Riffat R, Li B, Naz I, Badshah M, Ahmed S, Ali N (2019) Microbial population dynamics in temperature phased anaerobic digestion of municipal wastewater sludge. J Chem Technol Biotechnol 94:1816–1831. https://doi.org/10.1002/jctb.5955
Xiao BY, Zhang WZ, Yi H, Qin Y, Wu J, Liu JX, Li YY (2019) Biogas production by two-stage thermophilic anaerobic co-digestion of food waste and paper waste: effect of paper waste ratio. Renew Energ 132:1301–1309. https://doi.org/10.1016/j.renene.2018.09.030
Yucai L, Ning LI, Jinling G, Dachun G, Xiaofen W, Zongjun C (2016) Microbial community structure variation during the startup of culture enrichment under thermophilic condition inoculated with a mesophilic community for anaerobic digestion. Acta Sci Circumst 36:1986–1997. https://doi.org/10.13671/j.hjkxxb.2015.0648
Tian GL, Zhang WD, Dong MH, Yang B, Zhu R, Yin F, Zhao XL, Wang YX, Xiao W, Wang Q, Cui XL (2017) Metabolic pathway analysis based on high-throughput sequencing in a batch biogas production process. Energy 139:571–579. https://doi.org/10.1016/j.energy.2017.08.003
Xiao Z, Lin M, Fan J, Chen Y, Zhao C, Liu B (2018) Anaerobic digestion of spent mushroom substrate under thermophilic conditions: performance and microbial community analysis. Appl Microbiol Biotechnol 102:499–507. https://doi.org/10.1007/s00253-017-8578-9
Chen H, Chang S (2017) Impact of temperatures on microbial community structures of sewage sludge biological hydrolysis. Bioresour Technol 245:502–510
Sluiter A, Ruiz R, Scarlata C, Sluiter J, Templeton D (2005) Determination of extractives in biomass. National Renewable Energy Laboratory Technical Report 510:42619
Siming H, Jiqiang D, Xueni L, Xiaowei D, Fu X, Feifu L (2005) Optimized CTAB protocol for extracting the total DNA of ramie. Xibei Zhiwu Xuebao 25:2193–2197
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Martin M (2011) Cut adapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17:10–12
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Human MC, Petrosino JF, Knight R, Birren BW (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. https://doi.org/10.1101/gr.112730.110
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. https://doi.org/10.1038/nmeth.2604
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible in ARB. Appl Environ Microbiol 72:5069–5072
Gonzalez-Martinez A, Garcia-Ruiz MJ, Rodriguez-Sanchez A, Osorio F, Gonzalez-Lopez J (2016) Archaeal and bacterial community dynamics and bioprocess performance of a bench-scale two-stage anaerobic digester. Appl Microbiol Biotechnol 100:6013–6033. https://doi.org/10.1007/s00253-016-7393-z
Castellano-Hinojosa A, Armato C, Pozo C, Gonzalez-Martinez A, Gonzalez-Lopez J (2018) New concepts in anaerobic digestion processes: recent advances and biological aspects. Appl Microbiol Biotechnol 102:5065–5076. https://doi.org/10.1007/s00253-018-9039-9
Software canoco 4.5. http://www.canoco4.5.com/
Zheng Y, Zhao J, Xu F, Li Y (2014) Pretreatment of lignocellulosic biomass for enhanced biogas production. Prog Energ Combust 42(1):35–53. https://doi.org/10.1016/j.pecs.2014.01.001
Zhao Y, Xu C, Ai S, Wang H, Gao Y, Yan L, Mei Z, Wang W (2019) Biological pretreatment enhances the activity of functional microorganisms and the ability of methanogenesis during anaerobic digestion. Bioresour Technol 121660. https://doi.org/10.1016/j.biortech.2019.121660
Liu T, Zhou X, Li Z, Wang X, Sun J (2019) Effects of liquid digestate pretreatment on biogas production for anaerobic digestion of wheat straw. Bioresour Technol 280:345–351. https://doi.org/10.1016/j.biortech.2019.01.147
Lauwers J, Appels L, Thompson IP, Degrève J, Van Impe JF, Dewil R (2013) Mathematical modelling of anaerobic digestion of biomass and waste: power and limitations. Prog Energ Combust 39:383–402. https://doi.org/10.1016/j.pecs.2013.03.003
Bahmani MA, Shafiei M, Karimi K (2016) Anaerobic digestion as a pretreatment to enhance ethanol yield from lignocelluloses. Process Biochem 51:1256–1263. https://doi.org/10.1016/j.procbio.2016.05.012
Kumar R, Sharma RK, Singh AP (2017) Cellulose based grafted biosorbents - journey from lignocellulose biomass to toxic metal ions sorption applications - a review. J Mol Liq 232:62–93. https://doi.org/10.1016/j.molliq.2017.02.050
Li W, Khalid H, Zhu Z, Zhang R, Liu G, Chen C, Thorin E (2018) Methane production through anaerobic digestion: participation and digestion characteristics of cellulose, hemicellulose and lignin. Appl Energ 226:1219–1228. https://doi.org/10.1016/j.apenergy.2018.05.055
Pore SD, Shetty D, Arora P, Maheshwari S, Dhakephalkar PK (2016) Metagenome changes in the biogas producing community during anaerobic digestion of rice straw. Bioresour Technol 213:50–53. https://doi.org/10.1016/j.biortech.2016.03.045
Neshat SA, Mohammadi M, Najafpour GD, Lahijani P (2017) Anaerobic co-digestion of animal manures and lignocellulosic residues as a potent approach for sustainable biogas production. Renew Sust Energ Rev 79:308–322. https://doi.org/10.1016/j.rser.2017.05.137
Han S, Liu Y, Zhang S, Luo G (2016) Reactor performances and microbial communities of biogas reactors: effects of inoculum sources. Appl Microbiol Biotechnol 100:987–995. https://doi.org/10.1007/s00253-015-7062-7
Gieg LM, Davidova IA, Duncan KE, Suflita JM (2010) Methanogenesis, sulfate reduction and crude oil biodegradation in hot Alaskan oilfields. Environ Microbiol 12:3074–3086. https://doi.org/10.1111/j.1462-2920.2010.02282.x
Mbadinga SM, Li KP, Zhou L, Wang LY, Yang SZ, Liu JF, Gu JD, Mu BZ (2012) Analysis of alkane-dependent methanogenic community derived from production water of a high-temperature petroleum reservoir. Appl Microbiol Biotechnol 96:531–542. https://doi.org/10.1007/s00253-011-3828-8
Sun R, Zhou A, Jia J, Liang Q, Liu Q, Xing D, Ren N (2015) Characterization of methane production and microbial community shifts during waste activated sludge degradation in microbial electrolysis cells. Bioresour Technol 175:68–74. https://doi.org/10.1016/j.biortech.2014.10.052
Grabowski A, Tindall BJ, Bardin V, Blanchet D, Jeanthon C (2005) Petrimonas sulfuriphila gen. nov., sp nov., a mesophilic fermentative bacterium isolated from a biodegraded oil reservoir. Int J Syst Evol Microbiol 55:1113–1121. https://doi.org/10.1099/ijs.0.63426-0
Wang X, Zhu M, Li F, Zhang C, Zhu X (2018) Long-term effects of multi-walled carbon nanotubes on the performance and microbial community structures of an anaerobic granular sludge system. Appl Microbiol Biotechnol 102:9351–9361. https://doi.org/10.1007/s00253-018-9273-1
Leite AF, Janke L, Lv Z, Harms H, Richnow HH, Nikolausz M (2015) Improved monitoring of semi-continuous anaerobic digestion of sugarcane waste: effects of increasing organic loading rate on methanogenic community dynamics. Int J Mol Sci 16:23210–23226. https://doi.org/10.3390/ijms161023210
Zealand AM, Mei R, Papachristodoulou P, Roskilly AP, Liu WT, Graham DW (2018) Microbial community composition and diversity in rice straw digestion bioreactors with and without dairy manure. Appl Microbiol Biotechnol 102:8599–8612. https://doi.org/10.1007/s00253-018-9243-7
Maus I, Wibberg D, Winkler A, Pühler A, Schnürer A, Schlüter A (2016) Complete genome sequence of the methanogen Methanoculleus bourgensis BA1 isolated from a biogas reactor. Genome Announc 4:e00568–e00516. https://doi.org/10.1128/genomeA.00568-16
Kougias PG, Campanaro S, Treu L, Zhu X, Angelidaki I (2017) A novel archaeal species belonging to Methanoculleus genus identified via de-novo assembly and metagenomic binning process in biogas reactors. Anaerobe 46:23–32. https://doi.org/10.1016/j.anaerobe.2017.02.009
Schlüter A, Bekel T, Diaz NN, Dondrup M, Eichenlaub R, Gartemann KH, Krahn I, Krause L, Krömeke H, Kruse O, Mussgnug JH, Neuweger H, Niehaus K, Pühler A, Runte KJ, Szczepanowski R, Tauch A, Tilker A, Viehöver P, Goesmann A (2008) The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology. J Biotechnol 136:77–90. https://doi.org/10.1016/j.jbiotec.2008.05.008
Jaenicke S, Ander C, Bekel T, Bisdorf R, Droge M, Gartemann KH, Junemann S, Kaiser O, Krause L, Tille F, Zakrzewski M, Puhler A, Schluter A, Goesmann A (2011) Comparative and joint analysis of two metagenomic datasets from a biogas fermenter obtained by 454-pyrosequencing. PLoS One 6:e14519. https://doi.org/10.1371/journal.pone.0014519
Rademacher A, Zakrzewski M, Schlüter A, Schönberg M, Szczepanowski R, Goesmann A, Pühler A, Klocke M (2012) Characterization of microbial biofilms in a thermophilic biogas system by high-throughput metagenome sequencing. FEMS Microbiol Ecol 79:785–799. https://doi.org/10.1111/j.1574-6941.2011.01265.x
Zakrzewski M, Goesmann A, Jaenicke S, Junemann S, Eikmeyer F, Szczepanowski R, Al-Soud WA, Sorensen S, Puhler A, Schluter A (2012) Profiling of the metabolically active community from a production-scale biogas plant by means of high-throughput metatranscriptome sequencing. J Biotechnol 158:248–258. https://doi.org/10.1016/j.jbiotec.2012.01.020
Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D, Brown A, Allen N, Naylor J, Stange-Thomann N, DeArellano K, Johnson R, Linton L, McEwan P, McKernan K, Talamas J, Tirrell A, Ye W, Zimmer A, Barber RD, Cann I, Graham DE, Grahame DA, Guss AM, Hedderich R, Ingram-Smith C, Kuettner HC, Krzycki JA, Leigh JA, Li W, Liu J, Mukhopadhyay B, Reeve JN, Smith K, Springer TA, Umayam LA, White O, White RH, Conway De Macario E, Ferry JG, Jarrell KF, Jing H, Macario AJ, Paulsen I, Pritchett M, Sowers KR, Swanson RV, Zinder SH, Lander E, Metcalf WW, Birren B (2002) The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res 12:532–542. https://doi.org/10.1101/gr.223902
Fontana A, Patrone V, Puglisi E, Morelli L, Bassi D, Garuti M, Rossi L, Cappa F (2016) Effects of geographic area, feedstock, temperature, and operating time on microbial communities of six full-scale biogas plants. Bioresour Technol 218:980–990. https://doi.org/10.1016/j.biortech.2016.07.058
Funding
This work was financially supported by grants from the Natural Science Foundation of China (31370146), Fujian Agriculture and Forestry University International Cooperation and Exchange Project (No. KXG15001A), Sub Project of National Science and Technology Support Program (2014BAD15B01-6), and Key Research and Development Plan of Jiangxi Province (20171ACF60005).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lei, Y., Xie, C., Wang, X. et al. Thermophilic Anaerobic Digestion of Arundo donax cv. Lvzhou No. 1 for Biogas Production: Structure and Functional Analysis of Microbial Communities. Bioenerg. Res. 13, 866–877 (2020). https://doi.org/10.1007/s12155-020-10105-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-020-10105-y