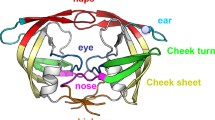
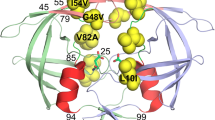
Abstract
HIV-1 protease (HIV-1PR) is an essential drug target in the treatment of patients infected with HIV-1. Mutations are found to arise in over 38 of 99 amino acid sites in this protein in response to drug therapy or natural selection, where many are found combinations that alter enzyme kinetics or inhibitor susceptibility without a clear structural mechanism. In efforts to understand how these mutations alter the flexibility and dynamics of HIV-1PR, we report the backbone 1H, 13C, and 15N chemical shift assignments for subtypes C, circulating recombinant form CRF01_AE and a multi-drug resistant variant MDR 769. These assignments are essential for future work aimed at characterizing backbone dynamics, exchange dynamics and dynamics of protein/substrate or protein/inhibitor interactions.


Similar content being viewed by others
References
Ashorn P, McQuade TJ, Thaisrivongs S, Tomasselli AG, Tarpley WG, Moss B (1991) An inhibitor of the protease blocks maturation of human and simian immunodeficiency viruses and spread of infection. Proc Natl Acad Sci USA 87:7472–7476
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfiefer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Freedberg DI, Ishima R, Jacob J, Wang YX, Kustanovich I, Louis JM, Torchia DA (2002) Rapid structural fluctuations of the free HIV protease flaps in solution: relationship to crystal structures and comparison with predictions of dynamics calculations. Protein Sci 11:221–232
Galiano L, Ding F, Veloro AM, Blackburn ME, Simmerling C, Fanucci GE (2009) Drug pressure selected mutations in HIV-1 protease alter flap conformations. J Am Chem Soc 131:430–431
Kear JL, Blackburn ME, Veloro AM, Dunn BM, Fanucci GE (2009) Subtype polymorphisms among HIV-1 protease variants confer altered flap conformations and flexibility. J Am Chem Soc 131:14650–14651
Longsdon BC, Vickrey JF, Martin P, Proteasa G, Koepke JI, Terlecky SR, Wawrzak Z, Winters MA, Merigan TC, Kovari LC (2004) Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity. J Virology 78:3123–3132
Mildner AM, Rothrock DJ, Leone JW, Bannow CA, Lull JM, Reardon IM, Sarcich JL, Howe WJ, Tomich CS, Smith CW (1994) The HIV-1 protease as enzyme and substrate: mutagenesis of autolysis sites and generation of a stable mutant with retained kinetic properties. Biochemistry 33:9405–9413
Prabu-Jeyabalan M, King NM, Nalivaika EA, Heilek-Snyder G, Cammack N, Schiffer CA (2006) Substrate envelope and drug resistance: crystal structure of RO1 in complex with wild-type human immunodeficiency virus type 1 protease. Antimicrob Agents Chemother 50:1518–1521
Sanches M, Krauchenco S, Martins NH, Gustchina A, Wlodawer A, Polikarpov I (2007) Structural characterization of B and non-B subtypes of HIV-protease: insights into the natural susceptibility to drug resistance development. J Mol Biol 369:1029–1040
Spinelli S, Liu QZ, Alzari PM, Hirel PH, Poljak RJ (1991) The three dimensional structure of the aspartyl protease from HIV-1 isolate BRU. Biochimie 73:1391–1396
Velazquez-Campoy A, Vega S, Freire E (2002) Amplification of the effects of drug resistance mutations by background polymorphisms in HIV-1 protease from African subtypes. Biochemistry 41:8613–8619
Weber IT, Agniswamy J (2009) HIV-1 protease: structural perspectives on drug resistance. Viruses 1:1110–1136
Acknowledgments
We acknowledge Dr. Jeff F. Ellena at UVA for his help with data collection. This work is supported by NSF MBC-0746533 (GEF), UF Department of Chemistry, UF Center for AIDS Research, and by NIH grant AI28571 (BMD). Acquisition of NMR data was supported through the National High Magnetic Field Laboratory (NHMFL) and obtained at the Advanced Magnetic Resonance Imaging and Spectroscopy (AMRIS) in the McKnight Brain Institute of the University of Florida.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, X., de Vera, I.M.S., Veloro, A.M. et al. Backbone 1H, 13C, and 15N chemical shift assignment for HIV-1 protease subtypes and multi-drug resistant variant MDR 769. Biomol NMR Assign 7, 199–202 (2013). https://doi.org/10.1007/s12104-012-9409-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-012-9409-7