Abstract
Calcium-binding protein 1 (CaBP1) regulates inositol 1,4,5-trisphosphate receptors (InsP3Rs) and a variety of voltage-gated Ca2+ channels in the brain. We report complete NMR chemical shift assignments of the Ca2+-saturated form of CaBP1 with Ca2+ bound at EF1, EF3 and EF4 (residues 1–167, BMRB no. 16862).
Similar content being viewed by others
Biological context
Neuronal calcium-binding proteins (CaBPs) belong to a subclass of the calmodulin (CaM) superfamily that regulates various Ca2+ channel targets in the brain and retina (Haeseleer et al. 2000). Multiple isoforms of CaBPs are localized in different neuronal cell types and perform specialized roles in signal transduction (Haeseleer et al. 2004). The CaBP1 isoform regulates the Ca2+ dependent activity of inositol 1,4,5-trisphosphate receptors (InsP3Rs) that serve as Ca2+ release channels on the endoplasmic reticulum membrane (Kasri et al. 2004). CaBP1 also regulates P/Q-type voltage-gated Ca2+ channels (Haeseleer et al. 2004), L-type channels, and the transient receptor potential channel, TRPC5 (Kinoshita-Kawada et al. 2005). CaBP1 contains four EF-hand motifs, but the second EF-hand (EF2) lacks critical residues required for high affinity Ca2+ binding (Wingard et al. 2005). Calcium-induced conformational changes in CaBP1 are important for promoting Ca2+-dependent regulation of InsP3Rs (Li et al. 2009) and other channel targets. Three-dimensional structures and NMR assignments are now known for CaBP1 in the Mg2+-bound, Ca2+-free state (Li et al. 2009) and for the protein with Mg2+ bound at EF1 and Ca2+ bound at EF3 and EF4 (Li et al. 2009). However, the structure is not yet known for Ca2+-saturated CaBP1 with Ca2+ bound at EF1, EF3 and EF4, which is a key signaling state for ion channel regulation. We report here the NMR assignments of CaBP1 with Ca2+-bound at EF1, EF3 and EF4, as a first step toward elucidating its atomic-level structure and regulatory mechanism.
Methods and experiments
Expression and purification of human CaBP1
All NMR experiments were performed on a small splice-variant of human s-CaBP1 referred to in this study as CaBP1. The recombinant CaBP1 protein was cloned into pET3b expression vector (Novagen) and over-produced in Escherichia coli strain BL21(DE3) as described previously (Wingard et al. 2005). 13C/15N-labeled protein expression was induced by the addition of 0.5 mM IPTG at 37°C in M9 minimal medium containing 15NH4Cl and [U-13C] glucose. Cells obtained from M9 cultures were disrupted by sonication. The cell lysate was centrifuged and the supernatant was loaded onto a Phenyl-Sepharose 4B column (Amersham Biosciences) and CaBP1 protein was purified as described (Wingard et al. 2005). Typically, 50 mg of purified protein was obtained from 1L culture. The protein identity and purity were confirmed by SDS–PAGE.
NMR spectroscopy
Samples for NMR analysis were prepared by dissolving 13C/15N-labeled CaBP1 (1 mM) in 0.3 ml of a 90/10% H2O/D2O or 100% D2O with 10 mM [2H11] Tris (pH 7.4), and 5 mM CaCl2. Under these conditions, CaBP1 contains Ca2+ bound at EF1, EF3 and EF4 (Wingard et al. 2005). All NMR spectra were recorded at 37°C on a Bruker Avance 600 or 800 MHz spectrometers with triple-resonance cryogenic probe. Backbone and side-chain chemical shift assignments were accomplished with 15N-HSQC, HNCO, CACBCONH, HNCACB, HBHACONH, 15N-HSQC-TOCSY, HCCH-TOCSY spectra. 1H chemical shift assignments of aromatic side chains were based on HBCBCGCDHD, 1H-1H TOCSY, 1H-1H NOESY spectra. Stereospecific assignments of valine and leucine methyl group were obtained by performing 13C-CT-HSQC experiments on protein samples with directed 13C labeling (Neri et al. 1989). NMR data were processed using NMR Pipe software package (Delaglio et al. 1995) and analyzed using SPARKY.
Assignments and data deposition
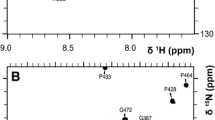
Figure 1 presents 15N HSQC spectrum of Ca2+-saturated CaBP1 (with Ca2+ bound at EF1, EF3 and EF4) to illustrate representative backbone resonance assignments. NMR assignments were based on 3D heteronuclear NMR experiments performed on 13C/15N-labeled CaBP1 (residues 1–167). The first 15 residues from the amino-terminus exhibited weak NMR signals that could not be assigned in our analysis. The remaining residues (16–167) exhibited strong NMR signals with uniform intensities, indicative of a well-defined three-dimensional protein structure in this region. More than 95% of the backbone resonances (1HN, 15N, 13Cα, 13Cβ, and 13CO), ~82% of aliphatic side chain resonances, and ~50% of aromatic side-chain resonances were assigned for residues in structured regions, including stereospecific assignment of valine and leucine methyl resonances. Three downfield shifted amide proton resonances at ~10.5 ppm are assigned to Gly40, Gly117 and Gly154 and verify that Ca2+ is bound at EF1, EF3 and EF4, consistent with our earlier Ca2+ binding analysis (Wingard et al. 2005). Chemical shift index analysis (Wishart and Sykes 1994) indicates the secondary structure of Ca2+-saturated CaBP1 is nearly identical to that observed for Mg2+-bound CaBP1 (Li et al. 2009). The chemical shift assignments (1H, 15N, 13C) of Ca2+-saturated CaBP1 have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under accession number 16862.
Two-dimensional 15N-1H HSQC spectrum of Ca2+-saturated CaBP1 recorded at 800-MHz 1H frequency. The protein sample (1 mM) was uniformly labeled with nitrogen-15 and was dissolved in 0.3 ml of a 90% H2O/10% [2H] H2O solution containing 10 mM [2H11] Tris (pH 7.4), 1 mM [2H10] dithiothreitol, and 5 mM CaCl2. Under these conditions, CaBP1 contains Ca2+ bound at EF1, EF3 and EF4 (Wingard et al. 2005)
Figure 2 presents amide chemical shift difference between residues of Ca2+-free and Ca2+-saturated CaBP1. The Ca2+-saturated CaBP1 chemical shifts of residues in EF2 show almost no effect of Ca2+ within experimental error, consistent with a lack of Ca2+ binding at EF2. The largest Ca2+-induced chemical shift changes are observed for residues in the Ca2+-binding loops of EF3 and EF4, consistent with high affinity Ca2+ binding at these sites. Much smaller Ca2+-induced chemical shift changes are observed for residues in the binding loop region of EF1. The relatively small Ca2+-induced chemical shift changes for residues in EF1 suggest that EF1 might remain in a closed conformation even in the Ca2+ bound state, in contrast to the Ca2+-induced open conformation observed previously for EF3 and EF4 (Li et al. 2009). The Ca2+-bound closed conformation for EF1 in CaBP1 is reminiscent of a Ca2+-bound closed conformation seen previously in cardiac troponin C (Wang et al. 2002). We propose that the closed conformation of EF1 in CaBP1 would prevent adventitious binding to protein targets like that shown for cardiac troponin C (Wang et al. 2002), and therefore might be functionally important for promoting highly specific target binding to CaBP1.
Amide chemical shift differences between Ca2+-free (Li et al. 2009) and Ca2+-saturated CaBP1 (this study). a shows ΔδN(ppm) = δN(Ca2+-free)—δN(Ca2+-bound) and b shows ΔδH(ppm) = δH(Ca2+-free)—δH (Ca2+-bound). Residues in the EF-hand binding loops display the largest chemical shift differences and are highlighted
References
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Haeseleer F, Sokal I, Verlinde CL, Erdjument H, Tempst P, Pronin AN, Benovic JL, Fariss RN, Palczewski K (2000) Five members of a novel Ca(2+)-binding protein (CABP) subfamily with similarity to calmodulin. J Biol Chem 275:1247–1260
Haeseleer F, Imanishi Y, Maeda T, Possin DE, Maeda A, Lee A, Reike F, Palczewski K (2004) Essential role of Ca2+-binding protein 4, a Cav1.4 channel regulator, in photoreceptor synaptic function. Nat Neurosci 7:1079–1087
Kasri NN, Holmes AM, Bultynck G, Parys JB, Bootman MD, Rietdorf K, Missiaen L, McDonald F, De Smedt H, Conway SJ, Holmes AB, Berridge MJ, Roderick HL (2004) Regulation of InsP3 receptor activity by neuronal Ca2+-binding proteins. EMBO J 23:312–321
Kinoshita-Kawada M, Tang J, Xiao R, Kaneko S, Foskett JK, Zhu MX (2005) Inhibition of TRPC5 channels by Ca2+-binding protein 1 in Xenopus oocytes. Pflugers Arch. 450:345–354
Li C, Chan J, Haeseleer F, Mikoshiba K, Palczewski K, Ikura M, Ames JB (2009) Structural Insights into Ca2+-dependent Regulation of Inositol 1, 4, 5-trisphosphate Receptors by CaBP1. J Biol Chem 284:2472–2481
Neri D, Szyperski T, Otting G, Senn H, Wuthrich K (1989) Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28:7510–7516
Wang X, Li MX, Sykes BD (2002) Structure of the regulatory N-domain of human cardiac troponin C in complex with human cardiac troponin I147–163 and bepridil. J Biol Chem 277:31124–31133
Wingard JN, Chan J, Bosanac I, Haeseleer F, Palczewski K, Ikura M, Ames JB (2005) Structural analysis of Mg2+ and Ca2+ binding to CaBP1, a neuron-specific regulator of calcium channels. J Biol Chem 280:37461–37470
Wishart DS, Sykes BD (1994) Chemical shifts as a tool for structure determination. Meth Enzymol 239:363–392
Acknowledgments
We thank Jerry Dallas for technical support and help with NMR experiments. Work supported by NIH grant (EY012347) to J.B.A.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Park, S., Li, C. & Ames, J.B. 1H, 15N, and 13C chemical shift assignments of calcium-binding protein 1 with Ca2+ bound at EF1, EF3 and EF4. Biomol NMR Assign 4, 159–161 (2010). https://doi.org/10.1007/s12104-010-9235-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-010-9235-8