Abstract
Objective
The common reference genes of choice in relative gene expression studies based on quantitative real time polymerase chain reaction, ACTB and B2M, were shown to be regulated differently in respect to tissue type. In this study, the stability of the selected housekeeping genes for normalizing the qPCR data were identified in the tumor and its adjacent tissues in invasive breast cancer, and the variability of their levels according to the stages and the histopathologic subtypes was analyzed.
Methods
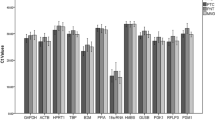
Four housekeeping genes: PUM1, RPL13A, B2M, and ACTB were analyzed in 99 surgically excised tissue specimens (50 tumor, 45 tumor adjacent and 4 normal breast tissues). Three of the most common softwares (GeNorm, NormFinder, and BestKeeper) were used for calculation purposes.
Results
When all of the tissue samples were included in analyses, PUM1 was the most stable gene according to calculations made with both NormFinder and BestKeeper; while PUM1/RPL13A combination was the most stable by GeNorm software. The PUM1 gene was also identified as the most stable gene among the four in all sample groups (in both Estrogen Receptor positive and Estrogen Receptor negative subgroups of invasive breast carcinoma and in normal breast tissue) according to calculations made using the NormFinder software.
Conclusion
While suggesting PUM1 is one of the most stable single gene and the PUM1/RPL13A pair as one of the best housekeeping genes for the normalization of expression studies in invasive breast tumor studies, it will be more practical to evaluate stability once more and decide upon the reference gene accordingly within the sample group itself.


Similar content being viewed by others
References
Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer. Classification, prognostication, and prediction. Lancet. 2011;378:1812–23.
Mocellin S, Rossi CR, Pilati P, Nitti D, Marincola FD. Quantitative real-time PCR: a powerful ally in cancer research. Trends Mol Med. 2003;9:189–95.
Yuan JS, Reed A, Chen F, Stewart CN. Statistical analysis of real-time PCR data. BMC Bioinform. 2006;7:85–96.
Jacques BK, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T, et al. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest. 2005;85:154–9.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–8.
Savonet V, Maenhaut C, Miot F, Pirson I. Pitfalls in the use of several “housekeeping” genes as standards for quantitation of mRNA: the example of thyroid cells. Anal Biochem. 1997;247(1):165–7.
Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, et al. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309(2):293–300.
Goidin D, Mamessier A, Staquet MJ, Schmitt D, Berthier-Vergnes O. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal Biochem. 2001;295(1):17–21.
El-Naggar AK, Mackay B, Sneige N, Batsakis JG. Stromal neoplasms of the breast: a comparative flow cytometric study. J Surg Oncol. 1990;44(3):151–6.
Freeman WM, Walker SJ, Vrana KE. Quantitative RT-PCR. pitfalls and potential. Biotechniques. 1999;26(1):112–22.
De Ferrari L, Aitken S. Mining housekeeping genes with a Naive Bayes classifier. BMC Genomics. 2006;7:277.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:1–11.
Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50.
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–15.
Çavuşoğlu AÇ, Saydam S, Alakavuklar M, Canda T, Kılıç Y, Harmancıoğlu O, et al. A pilot study for human tumor/DNA banking: returned more questions than answers. Med Oncol. 2008;25(4):471–3.
Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15:532–4.
Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64.
Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25(2):169–93.
Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6(4):279–84.
Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15(3):155–66.
Antonov J, Goldstein DR, Oberli A, Baltzer A, Pirotta M, Fleismann A, et al. Reliable gene expression measurements from degraded RNA by quantitative real-time PCR depend on short amplicons and a proper normalization. Lab Invest. 2005;85(8):1040–50.
Lyng MB, Laenkholm AV, Pallisgaard N, Ditzel HJ. Identification of genes for normalization of real-time RT-PCR data in breast carcinomas. BMC Cancer. 2008;8:20–30.
Majidzadeh-A K, Esmaeili R, Abdoli N. TFRC and ACTB as the best reference genes to quantify Urokinase Plasminogen Activator in breast cancer. BMC Res Notes. 2011;4:215.
Shah KN, Faridi JS. Estrogen, tamoxifen, and Akt modulate expression of putative housekeeping genes in breast cancer cells. J Steroid Biochem Mol Biol. 2011;125(3–5):219–25.
McNeill RE, Miller N, Kerin MJ. Evaluation and validation of candidate endogenous control genes for real-time quantitative PCR studies of breast cancer. BMC Mol Biol. 2007;27:107–19.
Tutt A, Wang A, Rowland C, Gillett C, Lau K, Chew K, et al. Risk estimation of distant metastasis in node-negative, estrogen receptor-positive breast cancer patients using an RT-PCR based prognostic expression signature. BMC Cancer. 2008;8:339.
Popovici V, Goldstein DR, Antonov J, Jaggi R, Delorenzi M, Wirapati P, et al. Selecting control genes for RT-QPCR using public microarray data. BMC Bioinformatics. 2009;10:42.
Khoshnoud R, He Q, Sylván M, Khoshnoud A, Ivarsson M, Fornander T, et al. The impact of RNA standardization and heterogeneous gene expression on the results of cDNA array of human breast carcinoma. Int J Mol Med. 2010;25(5):735–41.
Rienzo M, Schiano C, Casamassimi A, Grimaldi V, Infante T, Napoli C. Identification of valid reference housekeeping genes for gene expression analysis in tumor neovascularization studies. Clin Transl Oncol. 2013;15:211–8.
Zhang B. RefFinder Tool, Cotton EST Database, East Carolina, Department of Biology East Carolina University Greenville, NC, USA. 2012. http://www.leonxie.com/referencegene.php. Accessed April 2012.
Acknowledgments
The study was supported by the Scientific and Technological Research Council of Turkey (TÜBİTAK). The corresponding author, YK can be reached at +90-505-5985678 or yk@genodigm.com. The author AC was at the İzmir Training and Research Hospital, Department of Biochemistry at the time this project started. The authors would like to thank Dr. Kıvılcım Kılıç for her expertise in preparing the figures.
Conflict of interest
None declared.
Ethical standard
This study was performed under the ethical considerations of the Dokuz Eylül University Faculty of Medicine, Clinical and Laboratory Studies Ethical Council and with permission declared and numbered 03022006/03.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kılıç, Y., Çelebiler, A.Ç. & Sakızlı, M. Selecting housekeeping genes as references for the normalization of quantitative PCR data in breast cancer. Clin Transl Oncol 16, 184–190 (2014). https://doi.org/10.1007/s12094-013-1058-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-013-1058-5