Abstract
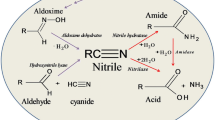
Nitrilases are commercial biocatalysts used for the synthesis of plastics, paints, fibers in the chemical industries, pharmaceutical drugs and herbicides for agricultural uses. Nitrilase hydrolyses the nitriles and dinitriles to their corresponding carboxylic acids and ammonia. They have a broad range of substrate specificities as well as enantio-, regio- and chemo-selective properties which make them useful for biotransformation of nitriles to important compounds because of which they are considered as ‘Green Catalysts’. Nitriles are widespread in nature and synthesized as a consequence of anthropogenic and biological activities. These are also present in certain plant species and are known to cause environmental pollution. Biotransformation using native organisms as catalysts tends to be insufficient since the enzyme of interest has very low amount in the total cellular protein, rate of reaction is slow along with the instability of enzymes. Therefore, to overcome these limitations, bioengineering offers an alternative approach to alter the properties of enzymes to enhance the applicability and stability. The present review highlights the aspects of producing the recombinant microorganisms and overexpressing the enzyme of interest for the enhanced stability at high temperatures, immobilization techniques, extremes of pH, organic solvents and hydrolysing dintriles to chiral compounds which may enhance the possibilities for creating specific enzymes for biotransformation.




Similar content being viewed by others
References
Sewell BT, Berman MN, Meyers PR, Jandhyala D, Benedik MJ (2003) The cyanide degrading nitrilase from Pseudomonas stutzeri AK61 is a two-fold symmetric, 14-subunit spiral. Structure 11(11):1413–1422. doi:10.1016/j.str.2003.10.005
Li C, Li Y, Cheng X, Feng L, Xi C, Zhang Y (2013) Immobilization of Rhodococcus rhodochrous BX2 (an acetonitrile-degrading bacterium) with biofilm-forming bacteria for wastewater treatment. Bioresour Technol 131:390–396. doi:10.1016/j.biortech.2012.12.140
Kumar V, Marin-Navarro J, Shukla P (2016) Thermostable microbial xylanases for pulp and paper industries: trends, applications and further perspectives. World J Microbiol Biotechnol 32(2):1–10. doi:10.1007/s11274-015-2005-0
Singh PK, Joseph J, Goyal S, Grover A, Shukla P (2016) Functional analysis of the binding model of microbial inulinases using docking and molecular dynamics simulation. J Mol Model 22(4):1–7. doi:10.1007/s00894-016-2935-y
Baweja M, Nain L, Kawarabayasi Y, Shukla P (2016) Current technological improvements in enzymes towards their biotechnological applications. Front Microbiol 7:965. doi:10.3389/fmicb.2016.00965
Gupta SK, Shukla P (2016) Advanced technologies for improved expression of recombinant proteins in bacteria: perspectives and applications. Crit Rev Biotechnol 36(6):1089–1098. doi:10.3109/07388551.2015.1084264
Baweja M, Singh PK, Shukla P (2015) Enzyme technology, functional proteomics and systems biology towards unraveling molecular basis for functionality and interactions in biotechnological processes. In: Shukla P (ed) Frontier discoveries and innovations in interdisciplinary microbiology. Springer, Berlin, pp 207–212. doi: 10.1007/978-81-322-2610-9
Kalia VC (2013) Quorum sensing inhibitors: an overview. Biotechnol Adv 31(2):224–245. doi:10.1016/j.biotechadv.2012
Kalia VC, Purohit HJ (2014) Quenching the quorum sensing system: potential antibacterial drug targets. Crit Rev Microbiol 37(2):121–140. doi:10.3109/1040841X.2010.532479
Kalia VC, Purohit HJ (2008) Microbial diversity and genomics in aid of bioenergy. J Ind Microbiol Biotechnol 35(5):403–419. doi:10.1007/s10295-007-0300-y
Nigam VK, Shukla P (2015) Enzyme based biosensors for detection of environmental pollutants—a review. J Microbiol Biotechnol 25(11):1773. doi:10.4014/jmb.1504.04010
Shukla P, Nigam V, Gupta R, Singh A, Kuhad RC (2013) Sustainable enzyme technology for environment: biosensors for monitoring of pollutants and toxic compounds. In: Kuhad RC, Singh A (eds) Biotechnology for environmental management and resource recovery. Springer India, New Delhi, pp 69–76. doi: 10.1007/978-81-322-0876-1
Singh PK, Imam J, Shukla P (2014) In-silico approach in bioremediation, microbial biodegradation and bioremediation. In: Das S (ed) Microbial biodegradation and bioremediation, 1st edn. Elsevier. doi: 10.1016/B978-0-12-800021-2.00027-3
Dubey KK, Kumar P, Singh PK, Shukla P (2014) Exploring prospects of mono-oxygenases based bio-catalyst in xenobiotics and their computational modeling. In: Das S (ed) Microbial biodegradation and bioremediation, 1st edn. Elsevier. doi: 10.1016/B978-0-12-800021-2.00027-3
Kumar V, Baweja M, Singh PK, Shukla P (2016) Recent developments in systems biology and metabolic engineering of plant-microbe interactions. Front Plant Sci 7:1421. doi:10.3389/fpls.2016.01421
Podar M, Eads JR, Richardson TH (2005) Evolution of a microbial nitrilase gene family: a comparative and environmental genomics study. BMC Evol Biol 5:42–46. doi:10.1186/1471-2148-5-42
Janowitz T, Kneifel H, Piotrowski M (2003) Identification and characterization of plant agmatine iminohydrolase, the last missing link in polyamine biosynthesis of plants. FEBS Lett 544:258–261. doi:10.1016/S0014-5793(03)00515-5
Baumann S, Sander A, Gurnon JR, Yanai-Balser GM, VanEtten JL, Piotrowski M (2007) Chlorella viruses contains genes encoding a complete polyamine biosynthetic pathway. Virology 360:209–217. doi:10.1016/j.virol.2006.10.010
Piotrowski M, Janowitz T, Kneifel H (2003) Plant C-N hydrolases and the identification of a plant N-carbamylputrescine amidohydrolase involved in polyamine biosynthesis. J Biol Chem 278:1708–1712. doi:10.1074/jbc.M205699200
Mueller P, Egorova K, Vorgias CE, Boutou E, Trauthwein H, Verseck S, Antranikian G (2006) Cloning, overexpression, and characterization of a thermoactive nitrilase from the hyperthermophilic archaeon Pyrococcus abyssi. Prot Exp Pur 47:672–681. doi:10.1016/j.pep.2006.01.006
Williamson DS, Dent KC, Brandon WW, Varsani A, Frederick J, Thuku RN, Cameron RA, Heerden JHV, Cowan DA, Sewell T (2010) Structural and biochemical characterization of a nitrilase from the thermophilic bacterium, Geobacillus pallidus RAPc8. Appl Microbiol Biotechnol 88:143–153. doi:10.1007/s00253-010-2734-9
Shao J, Li S, Zhang N, Cui X, Zhou X, Zhang G, Shen Q, Zhang R (2015) Analysis and cloning of the synthetic pathway of the phytohormone indole-3-acetic acid in the plant-beneficial Bacillus amyloliquefaciens SQR9. Microb Cell Fact 14(1):130. doi:10.1186/s12934-015-0323-4
Vejvoda V, Kubac D, Davidova A, Kaplan O, Sulc M, Sveda O, Chaloupkova R, Martinkova L (2010) Purification and characterization of nitrilase from Fusarium solani IMI196840. Pro Biochem 45(7):1115–1120. doi:10.1016/j.procbio.2010.03.033
Sharma NN, Sharma M, Kumar H, Bhalla TC (2006) Nocardia globerula NHB-2: bench scale production of nicotinic acid. Pro Biochem 41(9):2078–2081. doi:10.1016/j.procbio.2006.04.007
Fang S, An X, Liu H, Cheng Y, Hou N, Feng L, Huang X, Li C (2015) Enzymatic degradation of aliphatic nitriles by Rhodococcus rhodochrous BX2, a versatile nitrile-degrading bacterium. Biores Technol 185:24–28. doi:10.1016/j.biortech.2015.02.078
Bhalla TC, Miura A, Wakamoto A, Ohba Y, Furuhasi K (1992) Asymmteric hydrolysis of α-aminonitriles to optically active amino acids by a nitrilase of Rhodococcus rhodochrous PA-34. Appl Microbiol Biotechnol 37:184–190. doi:10.1007/BF00178168
Alamatawah QA, Cramp R, Cowan DA (1999) Characterization of an inducible nitrilase from a thermophilic Bacillus. Extremophiles 3:283–291
Kaplan O, Nikolau K, Pisvejcova CA, Martinkova L (2006) Hydrolysis of nitriles and amides by filamentous fungi. Enz Microb Technol 38:260–264. doi:10.1016/j.enzmictec.2005.07.022
Khandelwal AK, Nigam VK, Chaudhury B, Mohan MK, Ghosh P (2007) Optimization of nitrilase production from a new thermophilic isolate. J Chem Technol Biotechnol 82:646–651. doi:10.1002/jctb.1721
Vejvoda V, Kaplan O, Bezouska K, Pompach P, Sulc M, Cantarella M, Benada O, Uhnakova B, Rinagelova A, Wahl SL, Fischer L, Kren V, Martinkova L (2008) Purification and characterization of a nitrilase from Fusarium solani O1. J Mol Catal B Enzym 50:99–106. doi:10.1016/j.molcatb.2007.09.006
Kim J, Tiwari MK, Moon H, Jeya M, Ramu T, Oh D, Kim I, Lee J (2009) Identification and characterization of a novel nitrilase from Pseudomonas fluorescens Pf-5. Appl Microbiol Biotechnol 83:273–283. doi:10.1007/s00253-009-1862-6
Yang C, Wang X, Wei D (2011) A new nitrilase-producing strain named Rhodobacter sphaeroides LHS-305: biocatalytic characterization and substrate specificity. Appl Biochem Biotechnol 165:1556–1567. doi:10.1007/s12010-011-9375-z
Wang H, Li Guinan, Li Mingyang, Wei Dongzhi, Wang X (2014) A novel nitrilase from Rhodobacter sphaeroides LHS-305: cloning, heterologous expression and biochemical characterization. World J Microbiol Biotechnol 30:245–252. doi:10.1007/s11274-013-1445-7
Wang H, Gao W, Sun H, Chen L, Zhang L, Wang X, Wei D (2015) Protein engineering of a nitrilase from Burkholderia cenocepacia J2315 for efficient and enantioselective production of (r)-o-chloromandelic acid. Appl Environ Microbiol 81(24):8469–8477. doi:10.1128/AEM.02688-15
Dennett GV, Blamey JM (2016) A new thermophilic nitrilase from an antarctic hyperthermophillic microorganism. Front Bioeng Biotechnol 4:1–9. doi:10.3389/fbioe.2016.00005
Badoei-Dalfard A, Karami Z, Ramezani-pour N (2016) Nitrilase induction and characterization from a newly isolated Stenotrophomonas maltophilia AC21 and its application for bench scale production of nicotinic acid from 3-cyanopyridine. J Mol Catal B Enzym. doi:10.1016/j.molcatb.2016.11.019
Vergne-Vaxelaire C, Bordier F, Fossey A, Besnard-Gonnet M, Debard A, Mariage A, Salanoubat M (2013) Nitrilase activity screening on structurally diverse substrates: providing biocatalytic tools for organic synthesis. Adv Synth Catal 355(9):1763–1779. doi:10.1002/adsc.201201098
Brenner C (2002) Catalysis in the nitrilase superfamily. Curr Opinion Struc Biol 12:775–782. doi:10.1016/S0959-440X(02)00387-1
Winkler M, Kaplan O, Vejvoda V, Klempier N, Martinkova L (2009) Biocatalytic application of nitrilases from Fusarium solani O1 and Aspergillus niger K10. J Mol Catal B Enzym 59:243–247. doi:10.1016/j.molcatb.2008.06.012
Agarwal A, Nigam VK (2014) Nitrilase mediated conversion of indole-3-acetonitrile to indole-3-acetic acid. Biocatal Agric Biotechnol 3(4):351–357. doi:10.1016/j.bcab.2014.05.005
Raj J, Singh N, Prasad S, Seth A, Bhalla TC (2007) Bioconversion of benzonitrile to benzoic acid using free and entrapped cells of Nocardia globerula NHB-2. Acta Microbiol Immunol Hung 54:79–88. doi:10.1556/AMicr.54.2007.1.8
Kabaivanova L, Dimitrov P, Boyadzhieva I, Engibarov S, Dobreva E, Emanuilova E (2008) Nitrile degradation by free and immobilized cells of the thermophile Bacillus sp. UG-5B, isolated from polluted industrial water. World J Microbiol Biotechnol 24:2383–2388. doi:10.1007/s11274-008-9757-8
Nigam VK, Agarwal A, Sharma M, Ghosh P, Choudhury B (2009) Bioconversion of 3-cyanopyridine to nicotinic acid by a thermostable nitrilase. Res J Biotechnol 41:33–36
Chauhan S, Wu S, Blumerman S, Fallon RD, Gavagan JE, DiCosimo R, Payne MS (2003) Purification, cloning, sequencing and over-expression in Escherichia coli of a regioselective aliphatic nitrilase from Acidovorax facilis 72W. Appl Microbiol Biotechnol 61:118–122. doi:10.1007/s00253-002-1192-4
Wu S, Fogiel AJ, Petrillo KL, Jackson RE, Parker KN, DiCosimo R, Ben-Bassat A, O’Keefe DP, Payne MS (2008) Protein engineering of nitrilase for chemoenzymatic production of glycolic acid. Biotechnol Bioeng 99:717–720. doi:10.1002/bit.21643
Kaplan O, Bezouska K, Plihal O, Ettrich R, Kulik N, Vanek O, Kavan D, Benada O, Malandra A, Sveda O, Vesela A, Inagelova A, Slamova K, Cantarella M, Felsberg J, Duskova J, Dohnalek J, Kotik M, Kren V, Martinkova L (2011) Heterologous expression, purification and characterization of nitrilase from Aspergillus niger K10. BMC Biotechnol 11:1–15. doi:10.1186/1472-6750-11-2
Kaplan O, Bezouska K, Malandra A, Vesela A, Petrickova A, Felsberg J, Rinagelova A, Kren V, Martinkova L (2011) Genome mining for the discovery of new nitrilases in filamentous fungi. Biotechnol Lett 33:309–312. doi:10.1007/s10529-010-0421-7
Yusuf F, Rather IA, Jamwal U, Gandhi SG, Chaubey A (2015) Cloning and functional characterization of nitrilase from Fusarium proliferatum AUF-2 for detoxification of nitriles. Funct Integr Genom 15(4):413–424. doi:10.1007/s10142-014-0430-z
Yeom SJ, Kim HJ, Lee JK, Kim DE, Oh DH (2008) An amino acid at position 142 in nitrilase from Rhodococcus rhodochrous ATCC 33278 determines the substrate specificity for aliphatic and aromatic nitriles. Biochem J 415:401–407. doi:10.1042/BJ20080440
Demnerova K, Mackova M, Spevakova V, Beranova K, Kochankova L, Lovecka P, Ryslava E, Macek T (2005) Two approaches to biological decontamination of groundwater and soil polluted by aromatics characterization of microbial populations. Int Microbiol 8:205–211
Olaniran AO, Pillay D, Pillay B (2006) Biostimulation and bioaugmentation enhances aerobic biodegradation of dichloroethenes. Chemosphere 63:600–608. doi:10.1016/j.chemosphere.2005.08.027
Fantroussi El, Agathos S (2005) Is bioaugmentation a feasible strategy for pollutant removal and site remediation? Curr Opin Microbiol 8:268–275. doi:10.1016/j.mib.2005.04.011
Singh JS, Abhilash PC, Singh HB, Singh RP, Singh DP (2011) Genetically engineered bacteria: an emerging tool for environmental remediation and future perspectives. Gene 480(1–2):1–9. doi:10.1016/j.gene.2011.03.001
de Lorenzo V (2009) Recombinant bacteria for environmental release: what went wrong and what we have learnt from it. Clin Microbiol Infect 15:63–65. doi:10.1111/j.1469-0691.2008.02683.x
Schreiner U, Hecher B, Obrowsky S, Waich K, Klempier N, Steinkellner G, Gruber K, Rozzell JD, Glieder A, Winkler M (2010) Directed evolution of Alcaligenes faecalis nitrilase. Enzyme Microb Technol 47(4):140–146. doi:10.1016/j.enzmictec.2010.05.012
Sun H, Wang H, Gao W, Chen L, Kai W, We D (2015) Directed evolution of nitrilase PpL19 from Pseudomonas psychrotolerans L19 and identification of enantiocomplementary mutants toward mandelonitrile. Biochem Biophys Res Commun 468(4):820–825. doi:10.1016/j.bbrc.2015.11.038
Zhang XH, Liu ZQ, Xue YP, Zheng YG (2014) Activity improvement of a regioselective nitrilase from Acidovorax facilisand its application in the production of 1-(cyanocyclohexyl) acetic acid. Process Biochem 49(12):2141–2148. doi:10.1016/j.procbio.2014.08.018
Janowitz T, Trompetter I, Piotrowski M (2009) Evolution of nitrilases in glucosinolate-containing plants. Phytochemistry 70:1680–1686. doi:10.1016/j.phytochem.2009.07.028
Arfi T, Agarwal A, Nigam VK (2013) Bioproduction of nicotinic acid. Int J Pharm Technol 5(2):2622–2631
Piotrowski M (2008) Primary or secondary? Versatile nitrilases in plant metabolism. Phytochemistry 69:2655–2667. doi:10.1016/j.phytochem.2008.08.020
Gong JS, Lu ZM, Li H, Shi JS, Zhou ZM, Xu ZH (2012) Nitrilases in nitrile biocatalysis: recent progress and forthcoming research. Microb Cell Fact 11:1–18. doi:10.1186/1475-2859-11-142
Chen S, Gao H, Chen J, Wu J (2014) Surface modification of polyacrylonitrile fibre by nitrile hydratase from Corynebacterium nitrilophilus. Appl Biochem Biotechnol 174:2058–2066. doi:10.1007/s12010-014-1186-6
Acknowledgements
Tesnim Arfi duly acknowledges Senior Research Fellowship of Moulana Azad National Fellowship, U. G. C, Government of India (F1-17.1/2012-13/MANF-2012-13-MUS-JHA-9035), New Delhi, India for pursuing Ph.D.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nigam, V.K., Arfi, T., Kumar, V. et al. Bioengineering of Nitrilases Towards Its Use as Green Catalyst: Applications and Perspectives. Indian J Microbiol 57, 131–138 (2017). https://doi.org/10.1007/s12088-017-0645-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-017-0645-5