Abstract
Objective
To investigate the clinical, laboratory and genetic features of NAFLD patients based on MRI-PDFF in China.
Design
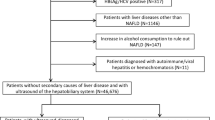
Patients with high ALT and with a diagnosis of fatty liver were included in this cross-sectional study. Fasting blood was collected to test biomarkers and SNPs. A total of 266 patients underwent MRI-PDFF and FibroScan examinations, and 38 underwent liver biopsy. Diagnostic models (decision tree, LASSO, and elastic net) were developed based on the diagnosis from MRI-PDFF reports.
Results
Approximately, 1/3 of the patients were found to have NASH and fibrosis. After quantifying liver steatosis by MRI-PDFF (healthy: n = 47; mild NAFLD: n = 136; moderate/severe NAFLD: n = 83; liver fat content (LFC): 3.6% vs. 8.7% vs. 19.0%), most biomarkers showed significant differences among the three groups, and patients without obesity were found to have a similar LFC as those with obesity (11.1% vs. 12.3%). Models including biomarkers showed strong diagnostic ability (accuracy: 0.80–0.91). Variant alleles of PNPLA3, HSD17B13 and MBOAT7 were identified as genetic risk factors causing higher LFC (8.7% vs. 12.3%; 11.0% vs. 14.5%; 8.5% vs. 10.2%, p < 0.05); those with the UQCC1 rs878639 variant allele showed lower LFC (10.4% vs. 8.4%; OR = 0.58, p < 0.05). Patients with more risk alleles had higher LFCs (8.1% vs. 10.7% vs. 11.6% vs. 14.5%).
Conclusions
Based on MRI-PDFF, a combination of several specific biomarkers can accurately predict disease status. When the effects of genes on liver steatosis were first quantified by MRI-PDFF, the UQCC1 rs878639 G allele was identified as a protective factor, and the MBOAT7 T allele was identified as a risk only among nonobese individuals.






Similar content being viewed by others
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
Abbreviations
- AC:
-
Abdominal circumference
- ALT:
-
Alanine transaminase
- AST:
-
Aspartate aminotransferase
- AUC:
-
Area under curve
- BMI:
-
Body mass index
- CAP:
-
Controlled attenuation parameter
- CK18:
-
Cytokeratin 18
- DBP:
-
Diastolic blood pressure
- FAST:
-
FibroScan-AST
- FGF21:
-
Fibroblast growth factor 21
- FLI:
-
Fatty liver index
- GGT:
-
Gamma-glutamyl transferase
- HC:
-
Hip circumference
- HCC:
-
Hepatocellular carcinoma
- HDL-C:
-
High-density lipoprotein cholesterol
- HSI:
-
Hepatocyte steatosis index
- IGF-1:
-
Insulin-like growth factor-1
- IL-6:
-
Interleukin-6
- IP-10:
-
Interferon-inducible protein 10
- IR:
-
Insulin resistance
- LDL-C:
-
Low-density lipoprotein cholesterol
- LFC:
-
Liver fat content
- LSM:
-
Liver stiffness measurement
- MAFLD:
-
Metabolic associated fatty liver disease
- MetS:
-
Metabolic syndrome
- MRI-PDFF:
-
Magnetic resonance imaging-derived proton density fat fraction
- NAFLD:
-
Non-alcoholic fatty liver disease
- NASH:
-
Non-alcoholic steatohepatitis
- OR:
-
Odds ratio
- SBP:
-
Systolic blood pressure
- T2DM:
-
Diabetes mellitus type 2
- TB:
-
Total bilirubin
- TBA:
-
Total bile acid
- TC:
-
Total cholesterol
- TG:
-
Triglyceride
- TIMP1:
-
Tissue inhibitor matrix metalloproteinase 1
- WC:
-
Waist circumference
References
Paik JM, Golabi P, Younossi Y, et al. Changes in the global burden of chronic liver diseases from 2012 to 2017: the growing impact of NAFLD. Hepatology. 2020;72:1605–1616
Eslam M, Fan JG, Mendez-Sanchez N. Non-alcoholic fatty liver disease in non-obese individuals: the impact of metabolic health. Lancet Gastroenterol Hepatol. 2020;5:713–715
Eslam M, Sanyal AJ, George J, et al. MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology. 2020;158:1999-2014.e1
Nan Y, An J, Bao J, et al. The Chinese society of hepatology position statement on the redefinition of fatty liver disease. J Hepatol. 2021;75:454–461
Bessone F, Razori MV, Roma MG. Molecular pathways of nonalcoholic fatty liver disease development and progression. Cell Mol Life Sci. 2019;76:99–128
Ekstedt M, Hagstrom H, Nasr P, et al. Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology. 2015;61:1547–1554
Leung JC, Loong TC, Wei JL, et al. Histological severity and clinical outcomes of nonalcoholic fatty liver disease in nonobese patients. Hepatology. 2017;65:54–64
Simon TG, Roelstraete B, Khalili H, et al. Mortality in biopsy-confirmed nonalcoholic fatty liver disease: results from a nationwide cohort. Gut. 2021;70:1375–1382
Davison BA, Harrison SA, Cotter G, et al. Suboptimal reliability of liver biopsy evaluation has implications for randomized clinical trials. J Hepatol. 2020;73:1322–1332
Tamaki N, Ajmera V, Loomba R. Non-invasive methods for imaging hepatic steatosis and their clinical importance in NAFLD. Nat Rev Endocrinol. 2022;18:55–66
Chen J, Duan S, Ma J, et al. MRI-determined liver fat correlates with risk of metabolic syndrome in patients with nonalcoholic fatty liver disease. Eur J Gastroenterol Hepatol. 2020;32:754–761
Shao C, Ye J, Li F, et al. Different predictors of steatosis and fibrosis severity among lean, overweight and obese patients with nonalcoholic fatty liver disease. Dig Liver Dis. 2019;51:1392–1399
Chen H, Zeng WK, Shi GZ, et al. Liver fat accumulation measured by high-speed T2-corrected multi-echo magnetic resonance spectroscopy can predict risk of cholelithiasis. World J Gastroenterol. 2020;26:4996–5007
Lee SW, Lee TY, Yang SS, et al. The association of non-alcoholic fatty liver disease and metabolic syndrome in a Chinese population. Hepatobiliary Pancreat Dis Int. 2017;16:176–180
Zheng X, Gong L, Luo R, et al. Serum uric acid and non-alcoholic fatty liver disease in non-obesity Chinese adults. Lipids Health Dis. 2017;16:202
Sun L, Wang Q, Liu M, et al. Albumin binding function is a novel biomarker for early liver damage and disease progression in non-alcoholic fatty liver disease. Endocrine. 2020;69:294–302
Dai J, Yi J, Zhang S, et al. Serum 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid is associated with lipid profiles and might protect against non-alcoholic fatty liver disease in Chinese individuals. J Diabetes Investig. 2019;10:793–800
Castera L, Friedrich-Rust M, Loomba R. Noninvasive assessment of liver disease in patients with nonalcoholic fatty liver disease. Gastroenterology. 2019;156:1264–81.e4
Alvarez-Sola G, Uriarte I, Latasa MU, et al. Fibroblast growth factor 15/19 (FGF15/19) protects from diet-induced hepatic steatosis: development of an FGF19-based chimeric molecule to promote fatty liver regeneration. Gut. 2017;66:1818–1828
Bedogni G, Bellentani S, Miglioli L, et al. The Fatty Liver Index: a simple and accurate predictor of hepatic steatosis in the general population. BMC Gastroenterol. 2006;6:33
Lee JH, Kim D, Kim HJ, et al. Hepatic steatosis index: a simple screening tool reflecting nonalcoholic fatty liver disease. Dig Liver Dis. 2010;42:503–508
Eslam M, George J. Genetic contributions to NAFLD: leveraging shared genetics to uncover systems biology. Nat Rev Gastroenterol Hepatol. 2020;17:40–52
Ji F, Liu Y, Hao JG, et al. KLB gene polymorphism is associated with obesity and non-alcoholic fatty liver disease in the Han Chinese. Aging (Albany NY). 2019;11:7847–7858
Liu Q, Liu SS, Zhao ZZ, et al. TRIB1 rs17321515 gene polymorphism increases the risk of coronary heart disease in general population and non-alcoholic fatty liver disease patients in Chinese Han population. Lipids Health Dis. 2019;18:165
Wang X, Liu Z, Peng Z, et al. The TM6SF2 rs58542926 T allele is significantly associated with non-alcoholic fatty liver disease in Chinese. J Hepatol. 2015;62:1438–1439
Meisner J, Albrechtsen A. Testing for Hardy-Weinberg equilibrium in structured populations using genotype or low-depth next generation sequencing data. Mol Ecol Resour. 2019;19:1144–1152
Kouvari M, Chrysohoou C, Skoumas J, et al. The presence of NAFLD influences the transition of metabolically healthy to metabolically unhealthy obesity and the ten-year cardiovascular disease risk: a population-based cohort study. Metabolism. 2021;128: 154893
Zhang J, Xu Q, Lai F, et al. Joint associations of metabolically healthy abdominal obesity and non-alcoholic fatty liver disease with prediabetes and diabetes in Chinese adults. BMJ Open Diabetes Res Care. 2021;9: e002362
Garcia-Carretero R, Vigil-Medina L, Barquero-Perez O, et al. Logistic LASSO and elastic net to characterize vitamin D deficiency in a hypertensive obese population. Metab Syndr Relat Disord. 2020;18:79–85
McEligot AJ, Poynor V, Sharma R, et al. Logistic LASSO regression for dietary intakes and breast cancer. Nutrients. 2020;12:2652
Zhou F, Zhou J, Wang W, et al. Unexpected rapid increase in the burden of NAFLD in China from 2008 to 2018: a systematic review and meta-analysis. Hepatology. 2019;70:1119–1133
Wieckowska A, Zein NN, Yerian LM, et al. In vivo assessment of liver cell apoptosis as a novel biomarker of disease severity in nonalcoholic fatty liver disease. Hepatology. 2006;44:27–33
Kliewer SA, Mangelsdorf DJ. Bile acids as hormones: the FXR-FGF15/19 pathway. Dig Dis. 2015;33:327–331
Wojcik M, Janus D, Dolezal-Oltarzewska K, et al. A decrease in fasting FGF19 levels is associated with the development of non-alcoholic fatty liver disease in obese adolescents. J Pediatr Endocrinol Metab. 2012;25:1089–1093
Schuster S, Cabrera D, Arrese M, et al. Triggering and resolution of inflammation in NASH. Nat Rev Gastroenterol Hepatol. 2018;15:349–364
Newsome PN, Sasso M, Deeks JJ, et al. FibroScan-AST (FAST) score for the non-invasive identification of patients with non-alcoholic steatohepatitis with significant activity and fibrosis: a prospective derivation and global validation study. Lancet Gastroenterol Hepatol. 2020;5:362–373
Pavlou M, Ambler G, Seaman SR, et al. How to develop a more accurate risk prediction model when there are few events. BMJ. 2015;351: h3868
Mi WF, Chen XM, Fan TT, et al. Identifying modifiable risk factors for relapse in patients with schizophrenia in China. Front Psychiatry. 2020;11: 574763
Ye Q, Zou B, Yeo YH, et al. Global prevalence, incidence, and outcomes of non-obese or lean non-alcoholic fatty liver disease: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2020;5:739–752
Wong VW, Wong GL, Chan RS, et al. Beneficial effects of lifestyle intervention in non-obese patients with non-alcoholic fatty liver disease. J Hepatol. 2018;69:1349–1356
Eslam M, Valenti L, Romeo S. Genetics and epigenetics of NAFLD and NASH: clinical impact. J Hepatol. 2018;68:268–279
Trepo E, Valenti L. Update on NAFLD genetics: from new variants to the clinic. J Hepatol. 2020;72:1196–1209
Tucker EJ, Wanschers BF, Szklarczyk R, et al. Mutations in the UQCC1-interacting protein, UQCC2, cause human complex III deficiency associated with perturbed cytochrome b protein expression. PLoS Genet. 2013;9: e1004034
Vetter K, Wurst W. Expression of a novel mouse gene “mbFZb” in distinct regions of the developing nervous system and the adult brain. Mech Dev. 2001;100:123–125
Neville MJ, Wittemans LBL, Pinnick KE, et al. Regional fat depot masses are influenced by protein-coding gene variants. PLoS ONE. 2019;14: e0217644
Sanna S, Jackson AU, Nagaraja R, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40:198–203
Brunt EM, Wong VW, Nobili V, et al. Nonalcoholic fatty liver disease. Nat Rev Dis Primers. 2015;1:15080
Acknowledgements
We would like to show our deepest gratitude to every author.
Funding
This study was supported by the National Natural Science Foundation of China.
Author information
Authors and Affiliations
Contributions
YA, DY, LG, and JN designed the study. LZ, XZ, YZ, HUZ, DZ, QD, HJ, HC, LX, HOZ, MJ, LC, ZW, and DW performed the imaging or biopsy examination. YA performed the experiments and analyzed the data. YA and DY wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
Aruhan Yang, Xiaoxue Zhu, Lei Zhang, Yingwen Zhang, Dezhi Zhang, Meishan Jin, Junqi Niu and Huimao Zhang declare that they have no conflict of interest.
Animal research
Not applicable.
Consent to participate
This study was approved by the Ethics Committee of the First Hospital of Jilin University (Ethical Approval Number: 19K096001), and written informed consent was obtained from all the participants.
Consent to publish
Every author consented to publish.
Clinical trials registration
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12072_2022_10355_MOESM6_ESM.pdf
Supplementary file6 Supplementary Fig. 1. Cross validation of LASSO for the penalty term. The λ values ranged from 0.00006 to 0.158323 with a minimal binomial deviance achieved at 0.0081 and a more stringent value of 0.0224. (PDF 416 KB)
12072_2022_10355_MOESM7_ESM.pdf
Supplementary file7 Supplementary Fig. 2. Correlation analysis between imaging results and histologic results. Spearman’s correlation analysis showed a stronger correlation of the LFC with steatosis and NAS score than FibroScan. (PDF 455 KB)
12072_2022_10355_MOESM8_ESM.pdf
Supplementary file8 Supplementary Fig. 3. (A) Selected variables in the classification tree models to differentiate NAFLD and healthy subjects. Supplementary Fig. 3. (B) Selected variables in classification tree models to differentiate mild NAFLD and moderate/severe NAFLD.The importance of variables in the classification tree models is shown, and only the top five most important variables are shown. (PDF 397 KB)
12072_2022_10355_MOESM9_ESM.pdf
Supplementary file9 Supplementary Fig. 4. (A) Selected variables in the classification tree models to differentiate NAFLD and healthy subjects among nonobese populations. Supplementary Fig. 4. (B) Selected variables in the classification tree models to differentiate mild NAFLD and moderate/severe NAFLD among the nonobese population. The importance of variables in the classification tree models among the non-obese group is shown. (PDF 400 KB)
12072_2022_10355_MOESM10_ESM.pdf
Supplementary file10 Supplementary Fig. 5. (A) Selected variables in the regression tree models to predict LFC. Supplementary Fig. 5. (B) Selected variables in the regression tree models to predict LFC among the nonobese population. The importance of variables in the regression tree models is shown. (PDF 398 KB)
Rights and permissions
About this article
Cite this article
Yang, A., Zhu, X., Zhang, L. et al. Non-invasive evaluation of NAFLD and the contribution of genes: an MRI-PDFF-based cross-sectional study. Hepatol Int 16, 1035–1051 (2022). https://doi.org/10.1007/s12072-022-10355-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12072-022-10355-2