Abstract
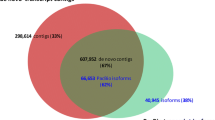
Sugarcane is the most important crop for sugar industry and raw material for bioethanol. Here we present a quantitative analysis of the gene content from publicly available sugarcane ESTs. The current sugarcane EST collection sampled orthologs for ~58 % of the closely-related sorghum proteome, suggesting that more than 10,000 sugarcane coding-genes remain undiscovered. Moreover the existence of more than 2,000 ncRNAs conserved between sugarcane and sorghum was revealed, among which over 500 are also detected in rice, supporting the existence of hundreds of conserved ncRNAs in grasses. New efforts towards sugarcane transcriptome sequencing were needed to sample the missing coding-genes as well as to expand the catalog of ncRNAs.

Similar content being viewed by others
References
Ben Amor B, Wirth S, Merchan F, Laporte P, d’Aubenton-Carafa Y, Hirsch J, Maizel A, Mallory A, Lucas A, Deragon JM, Vaucheret H, Thermes C, Crespi M (2009) Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res 19:57–69
Bower NI, Casu RE, Maclean DJ, Reverter A, Chapman SC, Manners JM (2005) Transcriptional response of sugarcane roots tomethyl jasmonate. Plant Sci 168:761–772
Carson D, Botha F (2002) Genes expressed in sugarcane maturing internodal tissue. Plant Cell Rep 20:1075–1081
Carson DL, Huckett BI, Botha FC (2002) Sugarcane ESTs differentially expressed in immature and maturing intermodal tissue. Plant Sci 162:289–300
Casu RE, Dimmock CM, Thomas M, Bower N, Knight D (2001) Genetic and expression profiling in sugarcane. Proc Int Soc Sugar Cane Technol 24:542–546
Casu RE, Grof CPL, Rae AL, McIntyre CL, Dimmock CM, Manners JM (2003) Identification of a novel sugar transporter homologue strongly expressed in maturing stem vascular tissues of sugarcane by expressed sequence tag and microarray analysis. Plant Mol Biol 52:371–386
De Lucia F, Dean C (2011) Long non-coding RNAs and chromatin regulation. Curr Opin Plant Biol 14(2):168–173
D’Hont A (2005) Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana. Cytogenet Genome Res 109:27–33
D’Hont A, Glaszmann JC (2001) Sugarcane genome analysis with molecular markers, a first decade of research. Proc Int Soc Sugar Cane Technol 24:556–559
Daniels J, Roach BT (1987) Taxonomy and evolution in sugarcane. In: Heinz D (ed) Sugarcane improvement through breeding. Elsevier Press, Amsterdam, pp 7–84
Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res 8:967–974
Garcia AA, Kido EA, Meza AN, Souza HM, Pinto LR, Pastina MM, Leite CS, Silva JA, Ulian EC, Figueira A et al (2006) Development of an integrated genetic map of a sugarcane (Saccharum spp.) commercial cross, based on a maximum-likelihood approach for estimation of linkage and linkage phases. Theor Appl Genet 112:298–314
Garsmeur O, Charron C, Bocs S, Jouffe V, Samain S, Couloux A, Droc G, Zini C, Glaszmann JC, Van Sluys MA et al (2011) High homologous gene conservation despite extreme autopolyploid redundancy in sugarcane. New Phytol 189:629–642
Goldemberg J (2006) The ethanol program in Brazil. Environ Res Lett 1:014008
Grivet L, D’Hont A, Roques D, Feldmann P, Lanaud C, Glaszmann JC (1996) RFLP mapping in cultivated sugarcane (Saccharum spp.): genome organization in a highly polyploid and aneuploid interspecific hybrid. Genetics 142:987–1000
Grivet L, Glaszmann JC, Vincentz M, da Silva F, Arruda P (2003) ESTs as a source for sequence polymorphism discovery in sugarcane: example of the Adh genes. Theor Appl Genet 106(2):190–197
Grivet L, Daniels C, Glaszmann JC, D’Hont A (2004) A review of recent molecular genetics evidence for sugarcane evolution and domestication. Ethnobot Res Appl 2:9–17
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Gupta V, Raghuvanshi S, Gupta A, Saini N, Gaur A, Khan MS, Gupta RS, Singh J, Duttamajumder SK, Srtivastava S et al (2010) The water-deficit stress- and red-rot-related genes in sugarcane. Funct Integr Genomics 10:207–214
Hoarau JY, Grivet L, Offmann B, Raboin LM, Diorflar JP, Payet J, Hellmann M, D’Hont A, Glaszmann JC (2002) Genetic dissection of a modern sugarcane cultivar (Saccharum spp.). II. Detection of QTLs for yield components. Theor Appl Genet 105:1027–1037
Jannoo N, Grivet L, Chantret N, Garsmeur O, Glaszmann J-C, Arruda P, D’Hont A (2007) Orthologous comparison in a gene-rich region among grasses reveals stability in the sugarcane polyploid genome. Plant J 50:574–585
Johnson C, Kasprzewska A, Tennessen K, Fernandes J, Nan GL, Walbot V, Sundaresan V, Vance V, Bowman LH (2009) Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res 19:1429–1440
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518
Lam E, Shine J, da Silva J, Lawton M, Bonos S, Calvino M, Carrer H, Silva-Filho MC, Glynn N, Helsel Z et al (2009) Improving sugarcane for biofuel: engineering for an even better feedstock. Glob Chang Biol Bioenergy 1:251–255
Ma HH, Schulze S, Lee S, Yang M, Mirkov E, Irvine J, Moore P, Paterson A (2004) An EST survey of the sugarcane transcriptome. Theor Appl Genet 108:851–863
Mattick JS (2001) Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep 2:986–991
Mattick JS (2005) The functional genomics of noncoding RNA. Science 309:1527–1528
Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15:R17–R29
Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJM (2009) RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21(3):367–376
Ming R, Liu SC, Lin YR, da Silva J, Wilson W, Braga D, van Deynze A, Wenslaff TF, Wu KK, Moore PH, Burnquist W, Sorrells ME, Irvine JE, Paterson AH (1998) Detailed alignment of saccharum and sorghum chromosomes: comparative organization of closely related diploid and polyploid genomes. Genetics 150:1663–1682
Menossi M, Silva-Filho MC, Vincentz M, Van-Sluys M, Souza GM (2008) Sugarcane functional genomics: gene discovery for agronomic trait development. Int J Plant Genomics 2008:458732
Mercer TR, Dinger ME, Mattick JS (2009) Long noncoding RNAs: insights into function. Nat Rev Genet 10:155–159
Moore PH (1995) Temporal and spatial regulation of sucrose accumulation in the sugarcane stem. Aust J Plant Physiol 22:661–679
Oliveira KM, Pinto LR, Marconi TG, Margarido GRA, Pastina MM, Teixeira LHM, Figueira AV, Ulian EC, Garcia AAF, Souza AP (2007) Functional integrated genetic linkage map based on EST-markers for a sugarcane (Saccharum spp.) commercial cross. Mol Breed 20:189–208
Ouyang S, Buell CR (2004) The TIGR plant repeat databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res 32:D360–D363
Pastina MM, Pinto LR, Oliveira KM, Souza KM, Garcia AAF (2010) Molecular mapping of complex traits. In: Henry (ed) Genetics, genomics and breeding of sugarcane. CRC Press, Science Publishers
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Piperidis G, Piperidis N, D’Hont A (2010) Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol Genet Genomics 284:65–73
Somerville C, Youngs H, Taylor C, Davis SC, Long SP (2010) Feedstocks for lignocellulosic biofuels. Science 329:790–792
Song X, Li P, Zhai J, Zhou M, Ma L, Liu B, Jeong DH, Nakano M, Cao S, Liu C, Chu C, Wang XJ, Green PJ, Meyers BC, Cao X (2011) Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J 69:462–474
Vettore AL, da Silva FR, Kemper EL, Souza GM, da Silva AM, Ferro MIs, Henrique-Silva F, Giglioti EA, Lemos MVF, Coutinho LL et al (2003) Analysis and functional annotation of an expressed sequence tag collection for tropical crop sugarcane. Genome Res 13:2725–2735
Yoshikawa M, Peragine A, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Development. 15; 19(18):2164–2175
Yu J et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296:79–92
Zanca AS, Vicentini R, Ortiz-Morea FA, Del Bem LEV, da Silva MJ, Vincentz M, Nogueira FTS (2010) Identification and expression analysis of microRNAs and targets in the biofuel crop sugarcane. BMC Plant Biol 10:260
Zhu QH, Wang MB (2012) Molecular functions of long non-coding RNAs in plants. Genes 3(1):176–190
Acknowledgments
This work was funded by grants 08/58031-0 (RV) and 08/52071-0 (MV) from FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) and LEVDB received a PhD scholarship from FAPESP (2008/09105-1).
Author information
Authors and Affiliations
Corresponding authors
Additional information
R. Vicentini and L. E. V. Del Bem are first authors.
Communicated by: Robert Henry
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
List of unique sampled sugarcane protein-coding orthologs from sorghum or rice. (XLS 1882 kb)
Table S2
List of sugarcane putative ncRNAs (XLS 266 kb)
Table S3
Sugarcane putative ncRNAs showing positive hits against the TIGR Plant Repeat Databases (XLS 17 kb)
Table S4
Sugarcane putative ncRNAs with repetitive and/or low complexity sequences as indentified by RepeatMasker software (XLS 40 kb)
File S1
Fastq file containing sRNAs sequences of 23 to 25 nucleotides showing perfect match against sugarcane putative ncRNAs. (FASTQ 5453 kb)
Figure S1
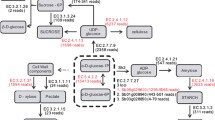
Schematic plot, using SeqMonk (http://www.bioinformatics.bbsrc.ac.uk/projects/seqmonk), of the 13 sugarcane ncRNAs most enriched in perfectly matched sRNAs (>1,000 sRNAs) showing examples of phase-distributed sRNAs. In the top graph, red and blue lines represent sRNAs mapped in plus or minus strand of the sugarcane CSC, respectively. The middle graph shows the quantification by heat-map of the mapped sRNAs. Finally, the bottom graph shows mVISTA (http://genome.lbl.gov/vista) conservation plot between sugarcane’s CSC and sorghum possible orthologs. (JPEG 143 kb)
Rights and permissions
About this article
Cite this article
Vicentini, R., Del Bem, L.E.V., Van Sluys, M.A. et al. Gene Content Analysis of Sugarcane Public ESTs Reveals Thousands of Missing Coding-Genes and an Unexpected Pool of Grasses Conserved ncRNAs. Tropical Plant Biol. 5, 199–205 (2012). https://doi.org/10.1007/s12042-012-9103-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-012-9103-z