Abstract
Parasites, and the diseases they cause, are important from an ecological and evolutionary perspective because they can negatively affect host fitness and can regulate host populations. Consequently, conservation biology has long recognized the vital role that parasites can play in the process of species endangerment and recovery. However, we are only beginning to understand how deeply parasites are embedded in ecological systems, and there is a growing recognition of the important ways in which parasites affect ecosystem structure and function. Thus, there is an urgent need to revisit how parasites are viewed from a conservation perspective and broaden the role that disease ecology plays in conservation-related research and outcomes. This review broadly focusses on the role that disease ecology can play in biological conservation. Our review specifically emphasizes on how the integration of tools and analytical approaches associated with both disease and molecular ecology can be leveraged to aid conservation biology. Our review first concentrates on disease-mediated extinctions and wildlife epidemics. We then focus on elucidating how host–parasite interactions has improved our understanding of the eco-evolutionary dynamics affecting hosts at the individual, population, community and ecosystem scales. We believe that the role of parasites as drivers and indicators of ecosystem health is especially an exciting area of research that has the potential to fundamentally alter our view of parasites and their role in biological conservation. The review concludes with a broad overview of the current and potential applications of modern genomic tools in disease ecology to aid biological conservation.
Similar content being viewed by others
Introduction
Parasites are the most ubiquitous, but possibly one of the least understood, members of global ecosystems. Ranging from microscopic viruses to tapeworms measuring over 10 m, parasites and the diseases they cause have long occupied a prominent place in social and scientific domains (see Appendix). However, despite our long fascination for parasites, we are only now beginning to understand how deeply they are embedded within the ecological systems. This is especially important in present times when human-mediated alterations in the environment from global climate change to local habitat fragmentation—have led to dramatic declines in biodiversity (Mace et al. 2012; Dirzo et al. 2014; Keith et al. 2015; Sato and Lindenmayer 2018) and have altered the structure and function of natural ecosystems. Such anthropogenic alterations can interact in complex ways to impact disease dynamics directly or indirectly in human and wildlife populations, creating a ‘geographic arena of pathogen emergence’ (Hoberg and Brooks 2015). Emerging and re-emerging diseases have been characterized as one of the most important challenges of our times, and it is increasingly becoming clear that managing parasites in natural populations depends upon viewing disease as an eco-evolutionary process.
Most diseases in natural populations, including recently emerging or re-emerging disease are a complex interplay of multiple host and parasite species as well as the broader ecological community (including nonhost species). This review focusses specifically on the role that parasites play in biological conservation, and comes a decade after the influential review by Smith et al. (2009) on the same topic. Consequently, here we primarily focus on the literature and/or concepts related to disease ecology that have been developed in the past decade. Also, given the substantial growth in the fields of genetics and genomics, our review also focusses specifically on how the integration of molecular and disease ecology can be leveraged to aid conservation biology. The first section focusses on the most apparent implications of parasites from a conservation perspective: disease-mediated extinctions and wildlife epidemics. The next sections take a step back and focus on elucidating how host–parasite interactions, specifically using genetic tools, has improved our understanding of the ecological and evolutionary dynamics affecting hosts at the individual, population, community and ecosystem scales (figure 1). We also discuss how parasites can serve as better ecological indicators at the host population and ecosystem scale. We believe that the role of parasites as drivers and indicators of ecosystem health is especially an exciting area of research that has the potential to fundamentally alter our view of parasites and their role in biological conservation. In the final section, we provide an overview of the recent advancements in molecular tools, especially high throughput DNA sequencing, that can be used to address broader questions in disease ecology such as enhanced disease surveillance, improved understanding of disease origin and spread, and spatio-temporal disease dynamics.
Schematic illustrating host, parasite and environmental factors influencing disease dynamics across scales of biological organization, at the individual, population, community and ecosystem scales. It is noteworthy that the effect of host and parasite factors on disease dynamics at each level is often not mutually exclusive and can range from individual to population and community/ecosystem levels.
Disease-mediated extinctions and epidemics in wildlife
There is an increasing recognition that wildlife diseases have important implications for biological conservation through their effects on biological diversity. Large-scale epidemics have occurred in natural populations leading to loss of biological diversity at local or global scales. Such epidemics have affected a wide variety of taxa, including the following. (i) Birds: one important group of emerging infectious diseases in birds are those attributed to avian haemosporidians (phylum Apicomplexa, order Haemosporida, genus Plasmodium and other related genera such as Haemoproteus and Leucocytozoon). Avian haemosporidians are a diverse group of vector-borne blood parasites that infect a wide array of bird taxa globally (Valkiŭnas 2005; Clark et al. 2014; Rivero and Gandon 2018). Large-scale mortalities in native wild birds have been well documented due to the accidental introduction of Plasmodium relictum and the vector Culex quinquefaciatus into island bird communities which had no coevolutionary history with these blood-borne parasites (e.g. Hawaii and New Zealand; Riper et al. 1986; Woodworth et al. 2005; Tompkins and Gleeson 2006). (ii) Amphibians: one of the most important emerging infectious diseases affecting amphibians is caused by the chytrid fungus, Batrachochytrium dendrobatidis. Chytridiomycosis has been linked to population declines, local extirpations and even global extinctions (Pounds et al. 2006; Skerratt et al. 2007). Initial reports indicated that the fungus likely originated in Africa (Weldon et al. 2004), but a more recent genomewide study suggests an Asian origin of B. dendrobatidis, and the pathogen was likely introduced globally with the expansion of commercial amphibian trade (O’Hanlon et al. 2018). Additionally, a newly discovered sister species, B. salamandrivorans has been associated with global population declines of salamanders (Martel et al. 2013; Yap et al. 2017). (3) Mammals: an important example of emerging infectious disease-causing catastrophic declines in mammalian populations is the white-nose syndrome (WNS) in bats. This disease is caused by the fungus Pseudogymnoascus destructans, a highly pathogenic fungus that colonizes the skin of hibernating bats (Blehert et al. 2009). The fungus was first identified in 2006 from New York and has caused local extirpations of many bats species, both endangered (e.g. Indiana bat, Myotis sodalist; Thogmartin et al. 2013) and formerly common ones (e.g., little brown bat, Myotis lucifugus; Frick et al. 2010) across North America.
Apart from the diseases mentioned above, a number of parasites have been implicated in population declines, including the rinderpest virus epidemic in wild ruminants in Kenya (Kock et al. 1999); the Ebola virus epidemic in Zaire that was responsible for the death of over 5000 gorillas (Gorilla gorilla; Bermejo et al. 2006); and the large-scale declines of North American bird populations due to West Nile virus (WNV; LaDeau et al. 2007).
In the face of large-scale epidemics, one major way in which molecular approaches can aid conservation is through the development of diagnostic tools to unambiguously identify parasites and quantify infection intensities (see section ‘Integrating genomics, disease, and conservation: current efforts and future directions’). These approaches have already been leveraged to develop diagnostic tools for a wide range of parasites and pathogens, including those that have been identified as being directly responsible for species endangerment, such as avian malaria (Richard et al. 2002) and WNV in birds (Kauffman et al. 2003), fungus causing WNS in bats (Lorch et al. 2010) and the chytrid fungus in amphibians (Rosenblum et al. 2013). However, animal extinctions caused solely by disease are likely to be rare because they will only occur under a relatively restricted set of ecological conditions. First, when a parasite is a generalist, capable of infecting more than one species, it can persist in a reservoir host (i.e., a host that is relatively unaffected by the parasite; De Castro and Bolker 2005). Indeed, most of the parasites cited above (i.e. B. dendrobatidis. P. relictum and the rinderpest virus) are generalist parasites infecting a broad diversity of hosts. Moreover, specific parasite strains may be more successful invaders into novel host communities as compared to others (e.g., P. relictum invasion in New Zealand; Ewen et al. 2012). The second mechanism that can lead to disease-mediated extinctions is frequency-dependent transmission (i.e. when transmission rate depends on the frequency of host contact, rather than host density; Boots and Sasaki 2003). Classic examples of frequency dependent transmission include vector-borne diseases (e.g., WNV and avian malaria), where high transmission rates can be maintained even at low host populations when the vector is efficient at finding their target species (Boots and Sasaki 2003). Consequently, from a conservation perspective, the most important effects of parasites are likely to be driven by their effects on other eco-evolutionary factors that directly or indirectly affect the process of species endangerment and recovery (see details below). The rest of our review focusses on how disease ecology, especially in conjunction with molecular ecology, can aid conservation by elucidating the varied eco-evolutionary interactions between parasites and their hosts.
Ecological and evolutionary dynamics
Parasites and the diseases they cause, negatively affect the organisms at multiple levels of biological organization: individuals, populations and species (Daszak et al. 2000; Smith et al. 2009; Tompkins et al. 2011) and alter the structure and function of ecological communities. At the individual level, parasites directly impact survival: mortality of infected animals is 2.65 times higher than uninfected ones (see meta-analysis by Robar et al. 2010). Parasites can reduce fitness through sub-lethal effects (e.g., impaired reproduction) or through indirect mechanisms (e.g., increased predation risk; Seppälä and Jokela 2008; Hatcher et al. 2014). Through their impacts on host fitness, parasites can also regulate host populations, limit host abundance, and lead to large-scale epidemic mortalities (see above). Further, parasites can alter the outcome of species interactions (e.g., competition, predation) and influence population and community structure (see section ‘Host-parasite transmission dynamics at the community and ecosystem level’). Finally, it is important to recognize that parasites are deeply embedded within our ecosystems and its effects can cascade through all trophic levels via alteration of food web structure, flow of energy and nutrients (Hatcher et al. 2012b; Wood and Johnson 2015).
Host–parasite transmission dynamics at the individual level
Numerous factors acting at the scale of the individual can impact parasite transmission dynamics. Here we focus on two factors, host genetic diversity and antiparasite defense, that modify the host susceptibility to infection.
Host genetic diversity
Parasites constitute a robust selective force on host populations because of their rapid evolutionary rate that can promote the maintenance of host genetic diversity (Ladle 1992). Many studies have indicated that host genetic diversity plays an important role in buffering host populations from diseases (King and Lively 2012). However, the relative importance of neutral versus adaptive genetic diversity has long been questioned in the field of conservation biology (e.g. Hughes 1991; Vrijenhoek and Leberg 1991). Low levels of neutral genetic diversity or high levels of inbreeding in natural populations are expected to increase host susceptibility to disease due to reduced adaptive potential. For instance, genetic diversity at neutral loci (measured as marker-based heterozygosity) has shown to be negatively associated with susceptibility to parasites in insects (e.g. Whitehorn et al. 2011), birds (e.g. Ortego et al. 2007a; Townsend et al. 2018), mammals (e.g. Mitchell et al. 2017; Rijks et al. 2008) and fish (e.g. Hedrick et al. 2001; Eszterbauer et al. 2015). Empirical evidence supports the perspective that inbred individuals generally tend to be more susceptible to parasites compared to outbred ones (Hedrick et al. 2001). Inbreeding has also been associated with increased disease severity. For example, American crows (Corvus brachyrhynchos) that died due to symptoms associated with WNV infections showed higher levels of inbreeding as compared to birds that died due to other causes (Townsend et al. 2009). Similarly, European treefrogs (Hyla arborea) from inbred populations died more quickly when exposed to B. dendrobatidis compared to those from outbred populations (Luquet et al. 2012).
However, the association between genetic diversity and parasitism is not universal, with some studies finding no association (Côté et al. 2005; Ortego et al. 2007b), or variable effects based on the strength of parasite-mediated selective pressures (Ruiz-López et al. 2012). Additionally, since inbreeding is negatively correlated with genetic diversity (e.g., marker-based heterozygosity), it is generally assumed that these metrics will affect parasite infection in opposite directions. Interestingly, Mitchell et al. (2017) found that while increased heterozygosity was associated with lower parasite loads in wild banded mongooses (Mungos mungo), there was no association with the pedigree-based inbreeding coefficient, likely due to linkage between genetic markers and genes influencing parasite burdens. Importantly, as genetic diversity increases, there is also a concomitant increase in the prevalence of rare genotypes in the population. The rarity of a particular host genotype could affect the infection risk if parasites tend to be coadapted to more common genotypes, as predicted by the Red Queen hypothesis (Lively et al. 1990; Lively and Dybdahl 2000; Decaestecker et al. 2007). However, a few studies have explicitly tested the relative effects of genotype rarity and heterozygosity on infection risk in wildlife. In an elegant study, Eastwood et al. (2017) showed that both heterozygosity and genotype rarity affect beak and feather disease virus infection risk in wild parrots (Platycercus elegans). Importantly, these authors show that heterozygosity reduced infection risk (but not infection load), while host genotype rarity decreased viral loads in infected individuals (but did not affect infection risk). Thus, in keeping with the Red Queen hypothesis, the more related an individual was to others within its population, the higher the infection risk. However, the metric used to measure ‘relatedness’ critically affected infection risk. Thus, the study found that nonspecific measures of relatedness (i.e. reduced overall heterozygosity) were associated with increased risk of infection (but did not affect the intensity of infection once a bird was infected). Alternatively, individual-specific measures of relatedness (i.e. genotype rarity) primarily affected parasite load, a pattern likely due to viral tracking of common host genotypes (i.e., the coevolution of viral genotypes with host susceptibility), assuming that risk of coinfection is low.
In contrast to neutral loci, functionally important loci (e.g. immune loci) can be more informative because genes associated with immune function generally seem to evolve more rapidly than other areas of the genome (Nielsen et al. 2005; Lazzaro and Clark 2012; Shultz and Sackton 2019). The major histocompatibility complex (MHC) is one of the most studied immune gene families in wild vertebrates due to its high variability and central role in adaptive immune response (Acevedo-Whitehouse and Cunningham 2006). Numerous studies have reported a negative relationship between MHC diversity and parasitism risk or disease severity in mammals (Sin et al. 2014; Caldwell and Siddle 2017), reptiles (Elbers and Taylor 2016), amphibians (Fu and Waldman 2017) and fish (Wegner et al. 2003). However, these patterns are not universal, with no effects of MHC diversity on infection risk in New Zealand passerine birds (Sutton et al. 2016) and giant pandas (Ailuropoda melanoleuca; Zhang et al. 2015).
MHC diversity could have important implications for community-level processes (e.g., interspecific competition). For example, the decline of red squirrels (Sciurus vulgaris) in the UK is likely due to apparent competition with the invasive eastern grey squirrels (Sciurus carolinensis) mediated by squirrelpox disease (see details in section on ‘Biological invasions’). Importantly, recent data have indicated that the increased susceptibility of red squirrels for the disease may be driven by MHC diversity because the red squirrel populations in the UK have much lower MHC diversity compared to populations in continental Europe (Ballingall et al. 2016). Studies have also revealed that selection tends to maintain high MHC diversity even in populations that have undergone recent, severe bottlenecks and have dramatically reduced neutral genetic diversity in numerous species, including Hume’s pheasant, Syrmaticus humiae (Chen et al. 2015); zebra finches, Taeniopygia guttata (Newhouse and Balakrishnan 2015); lowland leopard frog, Lithobates yavapaiensis (Savage and Zamudio 2016); alpine ibex, Capra ibex (Brambilla et al. 2018); black-spotted pond frog, Pelophylax nigromaculatus (Wang et al. 2017); European rabbits, Oryctolagus cuniculus (Schwensow et al. 2017). Strong balancing selection on MHC loci has also been shown in some populations, thus maintaining relatively uniform MHC diversities, despite population genetic differentiation at neutral markers (Niskanen et al. 2014). However, this study also revealed that in some parasites, individual MHC alleles may be more important than overall MHC diversity. Other studies have found similar patterns. For example, particular conformations of the MHC class II PBR appear to confer resistance to the chytrid fungus and these alleles are shared by parasite-resistant amphibian species globally (Fu and Waldman 2017). Similarly, in the case of avian malaria, individual MHC alleles or supertypes (functional clustering of alleles) seem to confer resistance to parasite infection in a wild great tit (Parus major) population (Sepil et al. 2013).
While MHC has been well studied, factors affecting MHC diversity in natural populations could be complex. For example, it has been hypothesized that mate choice should tend to maximize the offspring's MHC diversity to improve resistance to parasites (Von Schantz et al. 1996) and survival (Agbali et al. 2010). A meta-analysis across a wide variety of taxa suggests that the direct effects of sexual selection may outweigh those associated with parasites in maintaining MHC variation (Winternitz et al. 2013), and there is evidence that both diversity-based and dissimilarity-based mate choice act to maintain MHC diversity (Kamiya et al. 2014).
Another critical issue is that much of our understanding of wildlife immunogenetics has primarily focussed on the MHC family of genes, and there is a need for expanding the immune genes studied in wildlife. Many such candidate genes exist including chemokine, interleukin and toll-like receptors (TLRs), and interferon and tumour necrosis factor genes (Acevedo-Whitehouse and Cunningham 2006). Some of these genes have been investigated for their association with disease or immune competence in wildlife. For instance, Turner et al. (2012) reported that parasite-influenced selection maintains genetic diversity in cytokines, genes critical for initiating and mediating the immune response, in field voles (Microtus agrestis). A large-scale capture-mark-recapture study of wild bank voles (Myodes glareolus) showed that individual infection risk for Borrelia spp. infection was associated with specific genetic variants at the TLR gene (Tschirren et al. 2013). Further, a study by le Roex et al. (2013) revealed that three SNPs located in genes with predicted immune function were associated with bovine tuberculosis infection in the African buffalo (Syncerus caffer). Many studies in cervids also have shown that amino acid polymorphisms in the prion protein gene affects resistance to chronic wasting disease (Monello et al. 2017; Brandt et al. 2018). Finally, in one of the most comprehensive studies to date, Bateson et al. (2016) showed that in the endangered Attwater’s prairie-chicken (Tympanuchus cupido attwateri), innate (TLRs) and adaptive (MHC) immune systems, but not genomewide heterozygosity, affected the post-release survival of captive-bred birds.
Antiparasite defense
Traditional models of parasite virulence generally assume that parasites cause damage to their hosts because they acquire resources from their hosts to reproduce, and thus transmit infections to new hosts (Frank 1996). Thus, host–parasite interactions have traditionally been viewed as being antagonistic, with hosts maximizing fitness primarily by negatively impacting the parasite’s fitness through the development of resistance mechanisms. Such mechanisms either act to reduce infection risk (e.g., through behavioral mechanisms) or to reduce parasite burden once infected (e.g., immune-mediated killing; Råberg et al. 2009; De Roode and Lefèvre 2012; Medzhitov et al. 2012; Soares et al. 2017). However, it is increasingly being recognized that hosts can also minimize harm (fitness cost) inflicted at a given parasite burden through tolerance mechanisms (e.g., tissue repair; Ayres and Schneider 2012; Kutzer and Armitage 2016; Råberg et al. 2009, 2007; Schneider and Ayres 2008; Soares et al. 2017). The importance of parasite tolerance in plants has long been recognized (Caldwell et al. 1958; Pagán and García-Arenal 2018), but only recently have the animal ecologists started investigating the role of tolerance in shaping host–parasite interactions (above). Pioneering studies have revealed that variation in tolerance in insects could ameliorate infection-mediated reduction in survival (Ayres and Schneider 2009) or fecundity (Parker et al. 2014). While both resistance and tolerance improve host fitness in the face of parasite attack, they are expected to have very different outcomes. From an epidemiological perspective, resistance is expected to reduce parasite prevalence and select increased parasite virulence, while tolerance will have neutral or even positive effects on both prevalence and virulence (Schneider and Ayres 2008; Råberg et al. 2009; Ayres and Schneider 2012). From an evolutionary perspective, since tolerance does not directly reduce parasite fitness, antagonistic coevolution (i.e. Red Queen dynamics) is not expected in the case of tolerance, unlike in the case of resistance (Schneider and Ayres 2008; Råberg et al. 2009; Ayres and Schneider 2012; Kutzer and Armitage 2016). The relative investment in resistance versus tolerance can indeed affect disease dynamics at multiple scales of biological organization. For example, highly tolerant individuals are expected to be efficient at transmitting disease in a population (i.e. super-spreaders; Gopinath et al. 2014). Additionally, species that are tolerant to parasite infection may serve as reservoirs of infection and amplify the risk for vulnerable species. For example, the introduction of grey squirrels to Great Britain led to the decline of native red squirrel populations as grey squirrels were carriers of Parapoxvirus and were tolerant to infection (Rushton et al. 2000; Tompkins et al. 2002). Introduction of avian malaria in the Hawaiian Islands caused mass mortality and extinction of several native forest birds due to the virulent strain of P. relictum (GRW04) (Warner 1968; Atkinson et al. 2000). Empirical evidence suggests that lowland populations of Hawaii Amakihi (Hemignathus virens) that survived the infection had evolved tolerance as a defense strategy against avian malaria and may serve as reservoirs for local parasite populations (Atkinson et al. 2013). Vector-borne disease dynamics can also be affected by the evolution of tolerance in the vector. A recent study quantifying parasite resistance and tolerance in natural mosquito populations revealed that mosquitoes colonized from high transmission intensity areas had higher tolerance and lower resistance as compared to those colonized from low transmission intensity (Dharmarajan et al. 2019). Further, as tolerance was associated with increased vectorial capacity (i.e., the probability of a mosquito transmitting the parasite), this study showed that transmission intensity can impact vector evolution, which in turn can impact disease risk. Importantly, this study also showed that there was a tradeoff between resistance and tolerance as indicated by a significant negative relationship between these traits across the different natural populations. A negative relationship between resistance and tolerance has been reported previously (Stowe et al. 2000; Råberg et al. 2007), but such a relationship is not universal (Simms and Triplett 1994; Mauricio et al. 1997).
The view of the parasitic infections from a resistance-tolerance perspective necessarily delinks parasite infection risk and parasite burden from fitness consequences. For example, highly tolerant populations could have high parasite burdens with low fitness costs. Alternatively, populations with high resistance may have low parasite burdens but face costs associated with immunopathology. Consequently, the importance of eco-immunology (Hawley and Altizer 2011) and ecophysiology (Blaustein et al. 2012) both contribute to the progression of infection to disease. Methodological approaches to the measurement and analysis of resistance and tolerance in parasite populations are possible and continue to grow. However, there are multiple challenges in the interpretation of parasite load (e.g., the relative effects of increased infection risk versus reduced resistance), and the implications of parasite load on fitness (e.g., the confounding effects of immune-mediated vs. parasite-mediated pathology). Additionally, quantifying fitness and individual health in natural populations is a challenging issue. For instance, body-condition indices, a commonly used surrogate measure of fitness in ecological studies have been associated with some fundamental issues (e.g., many animals maintain body composition at homeostasis; Wilder et al. 2016). A recent meta-analysis reveals that while there is evidence for a negative relationship between condition indices and infection risk, the magnitude of this effect is relatively small and that there is evidence for publication bias towards negative relationships (Sánchez et al. 2018).
Taken together, while resistance versus tolerance are important host defense strategies in shaping parasite dynamics, hosts may not always evolve mechanisms to resist or tolerate parasite infections. Host behaviour and physiology can often play an important role in parasite transmission. Hosts can also evolve behavioural mechanisms to minimize or clear infection through sickness behaviours (e.g. coughing, fever, reduced social interactions and/or lethargy; Hart 1990) and/or self-medication (Ghai et al. 2015). In addition to infection induced changes in host behaviour which reduce parasite transmission, some parasites can manipulate host behaviour to increase their own transmission (i.e. adaptive manipulation; Moore 2002; Poulin 2010; Hughes et al. 2012). Some of the classic examples include hairworms causing crickets to jump into water where the worms reproduce (Thomas et al. 2002), brainworms inducing ants to bite into leaves to travel to the guts of ruminants (Moore 2002) and Toxoplasma gondii altering the behaviour of its rodent hosts by eliminating innate fear and inducing a fatal feline attraction that enables the parasite to reach its feline definitive host (Berdoy et al. 2000). Identifying the underlying physiological and/or developmental mechanisms for such parasite-induced behavioural changes is still challenging and is an active area of research (Hughes et al. 2012; Lafferty and Shaw 2013). Selection can act on parasitic traits to manipulate host behaviour if they increase parasite transmission, but the strength of this selection may depend on parasite life cycle (e.g. direct, vector-borne or trophic transmission) and/or host factors (e.g. spatial structure and social networks) at the population level.
Host–parasite transmission dynamics at the population level
Numerous mechanisms acting at the population scale can affect parasite transmission dynamics. Here, we focus on two of the most important factors, host spatial and social structure that can influence heterogeneity in contact rates, and thus parasite transmission risk.
Spatial structure
Molecular tools in conjunction with population/landscape genetic and phylogeographic analyses have been used to elucidate ecological and evolutionary processes across populations (Manel and Holderegger 2013; Petren 2013; Robin et al. 2015). The integration of such genetic methods with epidemiological information (e.g., parasite prevalence) has provided critical insights into disease transmission dynamics and epidemiological history of emerging infections (Archie et al. 2009). Levels of geneflow can help characterize the spatial spread of phenotypes of epidemiological importance (e.g., virulence, drug and pesticide resistance) in parasite and vector populations, and also help quantify parasite invasion risk (Schwabl et al. 2017). Thus, genetic data can aid biological conservation by providing insights into how ecological factors affect the functional connectivity of parasites, hosts and vectors at the individual or population scales. For example, fine-scale genetic data has been used to elucidate the spatio-temporal factors affecting how individual wildlife hosts sampled ticks from their environment (Dharmarajan et al. 2010, 2011). Specifically, this research produced empirical evidence for the aggregated transmission of parasites as ‘packets’ of related individuals, and such transmission dynamics, along with a subdivided mating system (i.e. mating at the host scale), could explain the deviations from Hardy–Weinberg equilibrium expectations, which is common in many parasites (Dharmarajan et al. 2011). Critically, such aggregated transmission dynamics in conjunction with host spatial structure could have critical epidemiological implications (e.g., the spread of recessive drug resistance genes; Dharmarajan 2015).
Underlying landscape features can critically alter host movement patterns and consequently influence the spatial spread of parasites (Biek and Real 2010; Kozakiewicz et al. 2018). Landscape genetics has been an extraordinarily powerful approach that has been used effectively to identify landscape features influencing the spatial spread of parasites, including chronic wasting disease prions due to movement of deer (Cullingham et al. 2011a; 2011b; Kelly et al. 2014; Brandt et al. 2018) and rabies virus due to movement of hosts like raccoons (Procyon lotor) and skunks (Mephitis mephitis; Heloise et al. 2012; Paquette et al. 2014). However, landscape features do not necessarily limit animal movement in all species and may facilitate parasite spread. For example, Deyoung et al. (2009) were unable to identify natural boundaries to geneflow and thus rabies spread for gray foxes (Urocyon cinereoargenteus). Landscape genetics approaches also form a powerful approach to quantify dispersal resistance/permeability surfaces for natural populations. For example, Bouyer et al. (2015) used microsatellite data to develop landscape resistance surfaces and thus identify candidate populations for tsetse fly (Glossina palpalis gambiensis) elimination.
Alternatively, the genetic structure of parasite populations can be used to elucidate characteristics of the host population that could not be obtained from host genetics alone (Dharmarajan et al. 2016). In the case of migratory birds, host genetics is a poor indicator of migratory routes since genetic recombination only occurs in the breeding grounds (summer) but not in nonbreeding (winter) grounds. However, critical insights into migratory strategies can be obtained through the genomic characterization of parasites, such as avian influenza virus in waterfowl (Hill et al. 2012; Newman et al. 2012).
Using phylogenetic approaches to analyse high-resolution parasite, genotype data can help detect and date historical changes in epidemiological patterns (Stadler and Bonhoeffer 2013), which can have implications for conservation. For example, genome resequencing of the chytrid fungus, shows that the fungus evolved in East Asia, and that the evolutionary history of this parasite likely predates its continent-scale movement (Rosenblum et al. 2013), which has likely been driven by commercial trade in amphibians (Fisher et al. 2012; O’Hanlon et al. 2018). Similarly, sequencing of the hemagglutinin gene of canine distemper virus isolates from over a period of four decades revealed that the virus emerged in the United States in the late 1800s, with subsequent diversification and global spread, likely due to uncontrolled animal trade and human transfer of pets (Panzera et al. 2015). Spatial spread of parasites is not necessarily associated with phylogenetic divergence. For example, in the case of Yersinia species, whole-genome sequencing has revealed a deep phylogenetic split between pathogenic strains and nonpathogenic lineages. However, all pathogenic Yersinia species do not share a recent common pathogenic ancestor, but instead they seem to have converged independently to acquire the same virulence determinants (Reuter et al. 2014).
For parasites that evolve rapidly, phylodynamics—an analytical method combining phylogenetics, epidemiology, population genetics, and immunology (Grenfell et al. 2004; Leventhal et al. 2012; Baele et al. 2017)—is a powerful approach that has been used to elucidate ecological (e.g., transmission bottlenecks; Volz et al. 2017) and evolutionary (virulence heritability; Vrancken et al. 2015) dynamics. RNA viruses are particularly well suited as markers for host ecology because they can spread via direct contact and have higher mutation rates compared to the host genome (Volz et al. 2013). Because of their relatively rapid evolutionary rate, viruses can reveal patterns of host movement and provide insight into patterns of disease distribution and spread that may not be apparent in the host genetic data, and thus provide a complementary tool for studying population dynamics of their hosts in ‘shallow’ time. For example, genetic patterns associated with feline immunodeficiency virus isolates from mountain lions (Puma concolor) in the Rocky Mountains of North America revealed pronounced spatial genetic structure providing information on the recent demographic history that was not evident from the host microsatellite data (Biek et al. 2006).
Social structure
Many animals show spatial heterogeneity in contact networks due to underlying ecological or environmental factors affecting levels of social structure. Social group size is generally driven by levels of competition for critical resources such as food (e.g., in golden jackal, Canis aureus; MacDonald 1979) or mates (e.g., in the striped mouse, Rhabdomys pumilio; Schradin et al. 2012). Aggregation of resources, such as supplemental feeding, can increase infectious disease transmission risk (Sorensen et al. 2013; Murray et al. 2016). Thus, bird aggregation around bird feeders during winter has been linked to outbreaks of mycoplasma conjunctivitis in North America (Dhondt et al. 2005). There is generally a positive association between group size and parasite infection risk (Griffin and Nunn 2012), and thus the elevated risk of parasitism could be a substantial cost associated with sociality. For example, gorillas in central Africa that clustered around seasonally fruiting trees were more affected by the outbreak of Ebola (97% mortality) compared to solitary individuals (77% mortality; Caillaud et al. 2006). At the population level, parasite transmission rates are expected to be higher in populations with low levels of social or spatial structure (i.e. smaller group sizes and more movements between groups; Craft 2015). For example, in the case of lions (Panthera leo), infection risk in animals living in prides is epidemiologically similar to homogeneous populations up to 20% larger (Caillaud et al. 2013).
Spatial aggregation of related individuals (i.e. kin structure) is a common form of social organization in wildlife (Altizer et al. 2003). Generally, we would expect increased levels of social interactions between related versus nonrelated individuals, and several studies have confirmed this pattern using epidemiological data. For instance, higher contact rates within family groups have been demonstrated in white-tailed deer (Odocoileus virginianus) owing to the fact that the deer infected by bovine tuberculosis (a parasite transmitted primarily by direct contact) are more closely related than noninfected deer (Blanchong et al. 2007). Similar findings have been reported in the case of chronic wasting disease in deer (Cullingham et al. 2011a; Magle et al. 2013). However, the effects of host relatedness on disease risk will depend up the spatial scale relevant for parasite transmission. For example, in raccoons, the infection risk associated with a directly transmitted parasite (canine distemper) was positively related to contact rates (i.e. familial structure) within spatially discrete habitat patches, while infection risk associated with an environmentally transmitted parasite (Leptospira spp.) was positively related to contact rates (i.e. geneflow) among the patches (Dharmarajan et al. 2012).
Social networks critically affect parasite transmission dynamics in wildlife (White et al. 2017), and consequently parasite networks can be used to provide valuable and often unique insights into contact/association patterns in natural populations. For example, in an interesting study, Chiyo et al. (2014) found that bacterial networks were not associated with social structure in African elephants (Loxodanta africana), but rather primarily driven by the habitat utilization patterns or individual host characteristics (i.e. sex and age). However, other studies have found that bacterial transmission networks strongly reflect underlying social structure of their hosts including sleepy lizards (Tiliqua rugosa; Bull et al. 2012) and giraffes (Giraffa camelopardalis; VanderWaal et al. 2014a).
Host–parasite transmission dynamics at the community and ecosystem levels
While disease ecology has traditionally focussed on dynamics at the individual and population scale, there is increasing evidence that community and ecosystem level process also critically affect transmission dynamics.
Species interactions
As in the case of individual species (see above), bacterial transmission networks have also been used to characterize patterns of species interactions in multi-species communities. For example, genetic analysis of Escherichia coli across 10 species of wild and domestic ungulates in Kenya revealed that the Grant’s gazelle (Gazella granti) showed the highest number of network connections in the community (VanderWaal et al. 2014b). However, the zebra (Equus burchelli), which tends to move longer distances than many other ungulates in this system, connected regions of the network that would otherwise have been poorly connected. Such studies at the community level have especially significant ramifications for emerging infectious diseases because characterizing disease emergence often entails a better understanding of when a parasite will spill over from one host species into another.
Ecological filters critically affect disease emergence because host switching is more likely for sympatric host species, and many emerging viruses in humans have zoonotic origins (e.g., SARS, corona and hanta viruses; Jones et al. 2008). Consequently, characterization of viruses harboured by potential wildlife reservoirs is a potential tool to predict or prevent disease emergence. For example, a metagenomic survey of viruses in the urine of the Old World fruit bat (Eidolon helvum) revealed that bats roosting close to humans harboured a wide variety of viruses, some of which are genetically related to known human parasites, highlighting the risk of zoonotic transmission (Baker et al. 2013). Whole-genome sequence data has also been used to characterize the parasite transmission at the wildlife–domestic animal interface, including the transmission of bovine tuberculosis from badgers (Meles meles) to cattle (Biek et al. 2012) and the spatial spread of Brucella abortus among livestock, bison (Bison bison) and elk (Cervus canadensis; Kamath et al. 2016).
Another novel way in which genetic tools (e.g., sequencing of mitochondrial genes) have been used to elucidate the potential for interspecific disease transmission is through the analyses of blood meals in disease-transmitting vectors such as mosquitoes (Crabtree et al. 2013), blackflies (Hellgren et al. 2008) and ticks (Gariepy et al. 2012). Such studies can be crucial for transmission dynamics. For instance, mosquito blood meal analysis combined with knowledge of host competence was used to identify which bird species were most important in the amplification of WNV (Hamer et al. 2009). This study predicted that the virus was primarily maintained by just three bird species (American robins, Turdus migratorius; blue jays, Cyanocitta cristata; house finches, Carpodacus mexicanus) which constituted a large proportion (35%, 17% and 15%, respectively) of hosts fed on by WNV-infectious Culex pipiens. From a conservation perspective, a particularly exciting avenue is that the vector blood meal analyses can be used as a tool for biodiversity prospecting (Bohmann et al. 2013; Lee et al. 2015; Schnell et al. 2015).
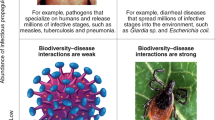
Biodiversity-disease relationship
The importance of host community structure on disease risk, especially zoonotic disease risk, in humans has been an area that has been especially contentious (Salkeld et al. 2013; Civitello et al. 2015). The dilution effect hypothesis posits host communities with high diversity are comprised of more hosts that are either refractory or less susceptible to parasites, and that such hosts ‘dilute’ the risk of disease in more competent hosts (Ostfeld and Keesing 2012; Johnson et al. 2015b). Consequently, this hypothesis predicts that reduced disease risk could be an ‘ecosystem service’ associated with biodiversity (Ostfeld and Logiudice 2003; Ostfeld and Keesing 2017). Other authors have argued that the ‘biodiversity protects against disease’ paradigm is Panglossian because it provides a questionable utilitarian justification for protecting biodiversity, and that, the specific composition of the host community critically decides whether biodiversity dilutes or amplifies disease risk (Randolph and Dobson 2012). The concept behind the dilution effect is not in itself new, and this has been proposed earlier in the ecological literature (e.g., the decoy effect; Chernin 1968), or has been effectively used to control vector-borne disease (e.g., zooprophylaxis; Hess and Hayes 1970). A recent meta-analysis has provided strong support for the dilution effect independent of host density, study design, and type and specialization of parasites (Civitello et al. 2015). However, there remains no definitive conclusion on the dilution effect debate (e.g. Ostfeld 2013; Randolph and Dobson 2012), and as in most ecological debates, the answer likely lies ‘somewhere in the middle’. For example, Keesing and Ostfeld (2012) mention that in two of the best case studies of the dilution effect (Lyme disease and WNV), the most competent host for the parasite (white-footed mice, Peromyscus leucopus and American robins, T. migratorius, respectively), are also the most resilient hosts that reach high abundance in degraded habitats. Basic ecological niche theory predicts that increasing levels of species diversity will favour the evolution of narrower niches to minimize niche overlap between species in a community. Consequently, generalist species are expected to dominate ecological communities with low species diversity and vice versa. Thus, the universality of the dilution effect could be driven by two potential mechanisms (Keesing and Ostfeld 2012): (i) successful parasites are those that have adapted to generalist hosts, thus areas with more generalist hosts (i.e. low diversity areas) will have higher infection risk; (ii) generalist hosts have evolved life history strategies that minimize allocation of resources to particular types of anti-parasite defense mechanisms (e.g. resistance), and thus areas with more generalist hosts (i.e. low diversity areas) will show higher infection risk.
Parasites and biological invasions
Invasive species are pervasive biotic agents that can impact native community structure and threaten global biodiversity, and the dual role of parasites as drivers of, as well as passengers responding to, altered community structure, is especially important from the perspective of biological invasions (Prenter et al. 2004; Dunn et al. 2012; Blackburn and Ewen 2017). Disease is as an important driver or consequence associated with nearly a quarter of species listed in the IUCN’s list of the world’s worst invaders (Hatcher et al. 2012a), and models reveal that parasites can affect the magnitude and rate of invasive species (Hilker et al. 2005; Coates et al. 2017). Parasites can drive the success (or failure) of biological invasions through three mechanisms: (i) the enemy release hypothesis (Keane and Crawley 2002) proposes that some invasive species may escape the adverse fitness effects (e.g., on survival or reproduction) associated with parasites in their native range and thrive better in their introduced range. This effect of parasites has been compared to the kryptonite effect (Hudson et al. 2006) because Superman gained super-powers on earth because he was freed from the effects of kryptonite. A meta-analysis found that parasite diversity and prevalence were higher in native versus exotic populations, indicating that enemy release could be common in biological invasions (Torchin et al. 2003). For example, the European green crabs (Carcinus maenas) that were introduced in North America carried only a subset of parasites from their native range, and consequently were larger in size and had better reproductive success (Torchin et al. 2001). (ii) Novel weapon hypothesis (Price et al. 1986) proposes that parasites can be cointroduced with invasive species that may spill over to native species. This can negatively impact the native species by influencing the competitive interactions between invasive and native species (i.e. through apparent-mediated or parasite-mediated competition). For instance, as mentioned earlier, the introduction of grey squirrels to Great Britain, which introduced Parapoxvirus and replaced native red squirrels is one of the best-documented cases of such parasite-mediated invasion. The Parapoxvirus caused a 20-fold greater mortality in red versus grey squirrels (Rushton et al. 2000; Tompkins et al. 2003). (iii) Biotic resistance hypothesis (Elton 1958) is similar but opposite to the novel weapons hypothesis, and proposes that introduced species could be negatively affected by resident parasite communities. Thus, generalist parasites could directly affect ecosystem stability through increased virulence in nonadapted alien hosts (i.e. Suicide King Dynamics). However, more broadly, it has been recognized that ecological diversity and complexity help maintain ecosystem resilience, and parasites may be important components of ecosystems because they constitute a large proportion of species diversity and dominate food web links in most ecosystems (see below). Consequently, it has been proposed that a healthy ecosystem is one that has high parasite species richness (Hudson et al. 2006).
While parasites may drive biological invasions, it is also possible that they are just passengers associated with biological invasions (Blackburn and Ewen 2017). The cointroduction of parasites with invasive species seems to be more common than previously recognized (Lymbery et al. 2014). Such cointroduced parasites are not only powerful tools for the reconstruction of invasion history (e.g. Blakeslee and Byers 2008) but may also alter the outcome of invasion (Lymbery et al. 2014). For example, a meta-analysis shows that parasite spillover from native to alien species is common, with native parasite species accounting for over 60% of the parasite fauna across a wide range of invasive taxa (Kelly et al. 2009). However, these authors also show that nonindigenous species can be highly competent hosts for such parasites and that there is evidence for spillback of these native parasites from the invasive to native species (Kelly et al. 2009). Consequently, parasites can affect and be affected by the structure of their host communities.
Parasites and ecosystem health
Myriad human-mediated environmental perturbations—habitat modification, chemical contamination and global climate change are increasingly impacting ecosystem structure and function and impairing the resilience of these systems to such perturbations. One way in which environmental perturbations affect human/wildlife populations is through altered disease dynamics, and consequently the emergence of novel pathogens or re-emergence of old ones (Rogalski et al. 2017). Such epidemiological endpoints are critical from the perspective of ecosystem health for three reasons. First, the sensitivity of disease dynamics to environmental modification makes parasites one of the best indicators of ecosystem health (see below). Second, altered disease dynamics can feedback on ecosystem health through its impact on various processes from community composition to nutrient cycling (Horwitz and Wilcox 2005; Johnson et al. 2015a; Preston et al. 2016). Third, the link between the altered structure and function of ecological systems and the health of humans, domestic animals and wildlife at individual and population scales is well recognized (Cable et al. 2017; Rogalski et al. 2017), and these concepts have particular importance from the perspective of emerging infectious disease (Bird and Mazet 2018). In this section, we primarily focus on how parasites form an integral component of ecosystems and alter food web structure, flow of energy and nutrients, and drive trophic cascades.
Parasites play an important role in maintaining the structure and functioning of food webs since they constitute a large proportion of total biomass in many ecosystems (Kuris et al. 2008; Preston et al. 2013). In one of the first empirical studies, Kuris et al. (2008) estimated that parasite biomass was similar to fishes and many invertebrate taxa; and exceeded that of avian predators in estuarine ecosystems. Similarly, it has been shown that the annual biomass of trematode larval stages could be up to 10-fold more than winter bird biomass and exceeded that of most aquatic invertebrate taxa (Preston et al. 2013). Besides their contribution to productivity in ecosystems, parasites can increase food web complexity by increasing connectivity, chain length and nestedness. For instance, studies have shown that parasites comprise a large proportion of trophic links (ranging from 29–78%) in aquatic ecosystems (Lafferty et al. 2006; Amundsen et al. 2009; Preston et al. 2014). The centrality of parasites in ecological networks is not surprising because, from a trophic perspective, parasites are predators (see Appendix), and thus, they can have cascading effects on many essential ecosystem processes such as decomposition (Sato et al. 2011) and grazing (Cleaveland et al. 2009). However, unlike true predators, parasites can also transfer energy within trophic levels (e.g., direct transmission of parasites between herbivores through grazing). Additionally, energy can move from parasites to true predators through direct (e.g., consumption of infected prey or parasite free-living stages; Johnson et al. 2010) or indirect (e.g., increasing susceptibility of intermediate hosts to predation through behavioural modification; Lefèvre et al. 2009) mechanisms.
Parasites, by definition, survive and reproduce by using resources obtained from their hosts. However, surprisingly a few empirical studies have highlighted the importance of parasites for cycling nutrients at the ecosystem scale (Vannatta and Minchella 2018). For example, frog tadpoles are dominant grazers in many stream ecosystems, and consequently high (98%) mortality of tadpoles due to chytrid fungus infections were associated with strong trophic cascades (increased algae and fine detritus biomass) and a 50% reduction in nitrogen cycling (Whiles et al. 2012). The study by Connelly et al. (2008) showed that chytrid-mediated local extirpation of over 90% of tadpoles led to similar large-scale trophic cascades and altered nutrient cycling in natural streams, including a 269% increase in chlorophyll, 220% increase in ash-free dry mass and 140% increase in inorganic sediments. Also, fungus-mediated mortality in alder trees (Alnus tenuifolia) has been shown to impact nitrogen cycling of flood plain ecosystems (Ruess et al. 2009). Other studies have found that parasite-mediated effects on nutrient cycling dynamics are similar/larger in magnitude than those attributed to better-characterized disturbances, such as climate change and air pollution (Lovett et al. 2010) or forest fires (Cobb et al. 2012).
As may be expected in the case of true predators, parasites can also drive trophic cascades by decreasing the abundance of herbivores and releasing producers from the pressures of herbivory (reviewed by Buck and Ripple, 2017). For instance, parasite-mediated mortality of rabbits due to Myxoma virus (Dobson and Crawley 1994), large ungulates due to rinderpest virus (Cleaveland et al. 2009), and amphibians due to chytrid fungus (Connelly et al. 2008; Whiles et al. 2012) have all led to cascading effects on producers. There are also examples where parasite-mediated mortality of true predators was associated with the release of herbivores from predatory pressures, including the mortality of red foxes (Vulpes vulpes) due to ectoparasitic mites (Lindström et al. 1994), and wolves (Canis lupus) due to canine parvovirus (Wilmers et al. 2006). Alternatively, because parasites are critically affected by host community structure, trophic cascades can feedback to alter disease dynamics. For example, long-term exclusion of herbivores has been associated with altered disease dynamics in grassland ecosystems (Keesing and Young 2014). A recent study (Weinstein et al. 2017) shows that the removal of large herbivores leads to increased rodent densities (due to reduced competition), which in turn leads to an increase in rodent-borne parasites. Consequently, parasites can be ‘drivers’ of trophic cascades or can simply be ‘passengers’ that respond to trophic cascades.
Parasites can serve as excellent indicators of ecosystem health. Historically, coal miners used canaries to detect low levels of carbon monoxide and methane gas (Holt and Miller 2011), and it has been proposed that parasites can act as ‘canaries’ to detect alterations in the health of ecological systems. One way in which parasites have been effectively used as bioindicators is with respect to accumulation of environmental contaminants, which may be driven by their higher trophic position as compared to their hosts. For example, concentrations of heavy metals in the tissue of certain parasites, like acanthocephalans of fish (Sures 2001; Nachev and Sures 2016; Sures et al. 2017b) and pinnipeds (McGrew et al. 2018) can be several magnitudes higher than those present in host tissues or environment. However, this pattern is not universally the case. For example, Tellez and Merchant (2015) found that while parasites (i.e. intestinal trematodes) showed higher concentrations of some heavy metals (e.g. arsenic and zinc), hosts (i.e., the American alligator, Alligator mississippiensis) were better indicators of others (e.g. cadmium and lead).
Parasites may also be effective indicators of ecosystem structure and function because they are likely sensitive to perturbations, including anthropogenic effects on ecological systems that affect host community structure, species interaction dynamics and food web topology (Marcogliese 2005). One area that has received a lot of attention is in testing the effects of specific anthropogenic modifications of the environment, such as pollution or land-use change (e.g., urbanization) on parasite infection risk or community structure. With respect to parasite infection risk (i.e. parasite loads), despite several decades of effort, there remain no general trends, and four influential meta-analysis have found that effects of contaminants on parasite loads could be positive, negative, or absent depending on the parasite and pollutant (Lafferty 1997; Sures 2008; Blanar et al. 2009; Vidal-Martínez et al. 2010). Alternatively, contaminants seem to have a relatively consistent negative effect on species richness (Blanar et al. 2009). For example, Calegaro-Marques and Amato (2014) found that helminth species richness in rufous-bellied thrushes (Turdus rufiventris) was inversely related to the degree of urbanization, and hypothesized that structures in urbanized landscapes (i.e. buildings) act to break up host–parasite interactions. Similarly, T. gondii genotype diversity was found to decrease in areas of human settlement (Jiang et al. 2018), and lead intoxication was found to reduce helminth species richness in mallards (Anas platyrhynchos; Prüter et al. 2018). It has also been proposed that the ratio between species richness of heteroxenous versus monoxenous parasites (i.e. parasites with complex versus direct life cycles, respectively) can be used as a measure of pollution impact (Pérez-del Olmo et al. 2007; Sures 2008; Sures et al. 2017b). Indeed, species richness of certain taxa (e.g., heteroxenous parasites like trematodes) has been used as a potential indicator of habitat restoration (Huspeni and Lafferty 2004; Morley and Lewis 2006; Sures et al. 2017a).
Integrating genomics, disease and conservation: current efforts and future directions
Recent advances in high-throughput sequencing technologies and concomitant development of bioinformatics tools have revolutionized the field of disease ecology. In this section, we illustrate how genomic tools have expanded the scope of wildlife disease research and refined our understanding of previously unexplored questions in disease ecology (Selbach et al. 2019). We highlight applications of genomic tools in five key research areas and hope to bridge the gap between genomics, disease and conservation to stimulate more interdisciplinary research (figure 2). First, applications of emerging molecular methodologies (e.g., whole-genome sequencing and metagenomics) have enhanced disease surveillance from individual hosts to ecosystem monitoring, improved management of disease outbreaks and can aid in biodiversity conservation. Second, phylogenomics can be used to infer the geographic origin of parasites, routes of parasite transmission, identify reservoir hosts, and provide more robust parasite phylogenies that can yield insights into the spatio-temporal patterns of parasite spread. Third, genomic approaches have allowed us to gain insights into fine-scale epidemiological patterns and contact tracing. Fourth, modern population genomic approaches (e.g. RAD-Seq) and genomewide scans provide a powerful platform to examine variation in disease susceptibility, and better understand host–parasite interactions. Fifth, transcriptomics or functional genomics can help us to better uncover genetic mechanisms underlying host resistance and/or tolerance and parasite virulence, elucidating host–parasite dynamics.
Applications of various high-throughput genomic tools (inner circle) to answer key questions in disease ecology (outer circle). Each genomic tool is represented by a different colour and colour gradients in the outer circle represent the multiple genomic approaches that can be used to answer a given question in the box. For instance, parasite origin and spread can be inferred using landscape genomics and/or phylogenomics.
Parasite detection and parasite genetic diversity
Molecular typing methods using traditional PCR techniques (e.g., genotyping or sequence analysis) have long been used to identify and characterize parasite infections as they are relatively easy to use and do not require any specialized lab setting or equipment. Quantitative PCR (qPCR) is another common diagnostic tool for sensitive and reliable identification of parasites, which also allows quantification of parasite load (Bell and Ranford-Cartwright 2002). More recently, the advent of droplet digital PCR (ddPCR) has advanced the detection and quantification of parasites, especially parasites with low intensity infections (Hindson et al. 2011; Pinheiro et al. 2012). It involves absolute end-point quantification of parasites and is more advantageous over qPCR, as it relies on calibration curves (generated from a sample with known concentration) to give relative quantification (Li et al. 2018). This technique has been successfully applied in accurate detection of several viral, bacterial diseases (King et al. 2017), and parasitic infections (Wilson et al. 2015; Koepfli et al. 2016; Ramírez et al. 2018).
Traditional PCR techniques are, however, limited in the amount of genetic information recovered as they use single or a few genetic markers. With reduced cost of high-throughput sequencing, rapid development in novel genomic approaches and bioinformatics, researchers can now access high resolution genomic level information for several parasites with relative ease at a fast pace, accurately identify multiple parasites simultaneously and assess parasite genetic diversity at both local and global spatial scales. Whole-genome sequencing and/or reduced genome technologies such as restriction-site associated DNA sequencing (RAD-SEQ) can provide thousands of markers (e.g. SNPs) for population genomics and/or phylogenomics and provide novel insights into disease transmission processes (Hupalo et al. 2015; Andrews et al. 2016). As more parasite genomes are being sequenced, huge amounts of genomic resources have become available for downstream applications such as comparative genomics, gene expression studies and improved insights into host–parasite interactions.
Most parasite genetic studies currently focus on one or a few related parasite species. However, more powerful genomic approaches such as, metagenomics can facilitate parasite discovery (Lipkin 2013), and help detect newly emerging pathogens or re-emerging pathogens in novel environments. By targeting a specific genetic region (e.g., 16S, 18S rDNA), metabarcoding or amplicon-based methods have improved our capability to characterize the structure and function of parasite communities in wildlife hosts (Bodewes et al. 2014; Tanaka et al. 2014; Bergner et al. 2019) and disease vectors (Ma et al. 2011). For example, ‘nemabiome’ sequencing, deep-amplicon sequencing of internal transcribed spacer 2 (ITS-2) rDNA has been used to characterize the diversity and composition of gastro-intestinal nematode parasite communities of grazing cattle in the mid-west USA (Avramenko et al. 2015) and in wild nonhuman primate populations in central Africa (Pafčo et al. 2018). In another recent study, a metabarcoding approach targeting 18S rRNA was used to characterize entire protozoan haemoparasite diversity from canine populations in Thailand (Huggins et al. 2019).
While metabarcoding has been recognized to have a great potential for parasite identification, many wildlife hosts often carry chronic infections, which makes it more difficult to obtain parasite genomic data due to the low abundance and/or intensity of parasites, without any prior enrichment strategy or when there is a high host contamination. One of the common approaches aimed at increasing the probability of capturing parasite specific DNA is depletion of host DNA. For instance, Flaherty et al. (2018) undertook a novel approach of restriction enzyme digestion of host DNA prior to enrichment and 18S rRNA amplicon sequencing of blood-borne parasites in human clinical samples. This method substantially reduced human reads and increased parasitic reads 5–10 folds relative to undigested samples. Another promising approach is target capture enrichment, which utilizes parasite-specific oligonucleotide probes or baits sequences to increase the proportion of parasite-specific genomic sequences in mixed species samples (Mamanova et al. 2010; Jones and Good 2016). For example, Lee et al. (2017) demonstrated potential use of target capture enrichment to obtain genomic sequences for multiple viral and bacterial pathogens from felids. The authors obtained up to 56 million-fold enrichment of pathogen DNA relative to the host DNA. As target capture enrichment uses probes developed from highly conserved sequences, this platform can also identify highly divergent and previously uncharacterized pathogens, in addition to the reference pathogens used to develop capture probes (Wylie et al. 2015; Barrow et al. 2018).
Another promising application of metagenomics has been to detect and characterize all microbial taxa within hosts and better understand the relationship between host microbiome and disease. Most eukaryotic organisms host many diverse microbial communities, which can be mutualistic, commensal or parasitic symbionts (Hooper et al. 2012). Taking this simplistic classification of host symbionts as discrete ecological entities rather than viewing them as a gradient along the parasitism–mutualism continuum has been challenging (Ewald 1987; Brown et al. 2012). Some of the key microbes that make up the resident microbiota can shift from a commensal to parasitic to mutualistic relationship in a context-dependent manner (e.g., changes in environmental conditions, tissue type, time and underlying biology of the interacting species; Leung and Poulin 2008). An extreme example of such a transition has been observed in Wolbachia spp., which evolved from being a parasite (reproductive manipulator) to a mutualist (enhanced host fecundity) in natural populations of Drosophila simulans over 20 years (Weeks et al. 2007). Thus, it is important to take a broad-spectrum approach when examining disease dynamics, as ecological and evolutionary interactions between symbionts, both beneficial and detrimental to hosts can influence the outcome of parasitic infections. Recently, Bass et al. (2019) has proposed the ‘pathobiome’ concept to encompass this diverse and complex relationship among the host-associated organisms to better understand the diseases of plants and animals.
High-throughput sequencing approaches such as metagenomics and metabarcoding have made it more feasible to characterize the host resident microbiota and its influence on host–parasitic interactions. A recent meta-analysis revealed disease associated shifts in human host microbiome, with both disease-specific and shared responses of gut microbial communities to multiple diseases (Duvallet et al. 2017). Shotgun metagenomics can also be used to investigate microbial communities in disease vectors carrying important human/wildlife vector-borne pathogens (ticks, Carpi et al. 2011; mosquitoes, Mancini et al. 2018; triatomines, Rodríguez-Ruano et al. 2018). Such information can be leveraged to advance our understanding of host–symbiont, symbiont–symbiont interactions from the perspective of infectious disease risk and developing novel strategies for disease control (see reviews for mosquitoes, Scolari et al. 2019; sand flies, Telleria et al. 2018, triatomines, Teotônio et al. 2019). Interestingly, Wolbachia spp., a bacterial endosymbiont found in Drosophila fruit flies that suppresses RNA viruses has been transfected in Aedes aegypti mosquitoes to combat dengue in several wild populations (reviewed in Dorigatti et al. 2018).
Host microbiome plays an important role in shaping the host immune system and can contribute to host defense against parasites (Chiu et al. 2017). A better understanding of interactions between host microbiome and parasitic infections can also aid in development of new therapeutic approaches for emerging/re-emerging diseases. The targeting of skin microbiota has gained momentum as an effective strategy against fungal pathogens. In the case of amphibian chytridiomycosis, skin microbiome is suggested to play an important role in host resistance and immune function (Bletz et al. 2013; Rebollar et al. 2016). Experimental studies have demonstrated that augmenting an antifungal bacterial species, Janthinobacterium lividum, to the skins of some amphibians can provide protective immunity against chytrid pathogen B. dendrobatidis and reduce morbidity and mortality of infected amphibians (Harris et al. 2009). In another study, composition of skin microbiota rather than treatment with antifungal bacteria improved the ability of Panamanian golden frogs (Atelopus zeteki) to clear B. dendrobatidis infection and ~30% of the infected individuals survived B. dendrobatidis exposure (Becker et al. 2015). Similarly, in bats susceptible to WNS, caused by the fungus P. destructans, treatment with symbiotic bacteria (Pseudomonas spp.) seems to be a promising tool to protect bats from WNS (Hoyt et al. 2015, 2019).
Taken together, these examples highlight the burgeoning potential of how host microbiome data has been invaluable for vector control strategies, tackling lethal emerging infectious diseases and improving host health and conservation. In addition to the importance of host microbiome, microbiome of parasites is important in understanding their biology and management of diseases. Despite the global concerted effort to characterize the animal microbiome through the Human Microbiome Project (Turnbaugh et al. 2007), or the Earth Microbiome Project (Gilbert et al. 2014), a similar initiative to characterize parasite microbiomes has only recently begun (Dheilly et al. 2017, 2019).
Moving up from within hosts to ecosystem scales, environmental DNA (eDNA) tools coupled with metagenomics can facilitate a more in-depth investigation of diversity of parasites and characterize all parasite taxa simultaneously from bulk environmental samples (e.g. water, soil, feces). In comparison to traditional field surveys and parasitic morphological identification approaches, eDNA approaches are a powerful, noninvasive approach (see Bass et al. 2015 for review) and particularly useful for detecting parasites with complex life cycles, which tend to shed transmission stages into the environment (Sengupta et al. 2019). Although, eDNA approaches have largely been used to detect the presence/absence of free-living species and biodiversity assessments (Taberlet et al. 2012; Bohmann et al. 2014; Thomsen and Willerslev 2015), they show promising potential in early detection of parasites, spread of invasive parasites, identifying reservoir host populations and quantifying disease risk at ecosystem scales. Until now, eDNA techniques have been successfully employed to detect amphibian pathogens from aquatic ecosystems, such as ranavirus (Miaud et al. 2019; Vilaça et al. 2019), the trematode Ribeiroia ondatrae (Huver et al. 2015) and the fungus B. dendrobatidis (Kamoroff and Goldberg 2017). Moreover, eDNA metabarcoding has also been used in early detection of disease vectors such as dipteran insects (Schneider et al. 2016) and phlebotamine sand flies (Kocher et al. 2017), which are known to transmit numerous diseases affecting human and wildlife populations.
It is, however, important to recognize that while metagenomics and/or metabarcoding approaches are promising tools for characterization of parasites from host (e.g. microbiome) or environment (e.g. eDNA), its applications are still limited. In metagenomics studies, accurate parasite detection relies on the availability of comprehensive reference databases that may limit the number of parasite species and taxonomic groups detected. We still lack high quality, well-annotated reference genomes for several parasite species, especially wildlife parasites. It could also be difficult to identify some eukaryotic parasites due to its dependency on an appropriate DNA marker for metabarcoding studies. For instance, conserved genetic loci for a eukaryotic parasite may be also be conserved across its eukaryotic host and may lead to disproportionately low amplification of parasite DNA.
Parasite origin and spread
Phylogeographic analysis of pathogen genomes using metagenomics and/or whole-genome sequencing is a powerful approach for identifying putative origins and examining evolutionary relationships among parasites (O’Hanlon et al. 2018). For instance, whole-genome sequencing of bat White-nose fungus supports the recent introduction of P. destructans to North America from Europe rather than Asia and subsequent loss of genetic diversity among the isolates from North America (Drees et al. 2017). In the case of raccoon rabies virus (RRV), which has largely been restricted to eastern North America but has recently been reported from Canada (Stevenson et al. 2016), whole-genome sequencing of RRV recovered multiple introduction events or backflow between US and Canada that resulted in multiple rabies outbreaks near the US–Canada border (Trewby et al. 2017).
Parasite genomic data can also aid in examining spatio-temporal patterns of disease spread, identifying transmission routes and improve prediction of emerging infectious disease risk (Faria et al. 2011; Dellicour et al. 2016). For example, viral RNA-seq and whole-genome sequence data have been effectively used to study the spread and transmission patterns of WNV in North America (Swetnam et al. 2018) and the spatial dynamics of the rabies virus in Africa (Brunker et al. 2018). Another novel application of pathogen genomics has been in identifying reservoir hosts. Babayan et al. (2018) analysed viral genome sequences and used a machine learning framework to predict reservoir hosts and/or arthropod vectors for 12 different RNA viral groups based on host-associated genomic biases.
Fine scale transmission dynamics: contact networks
At a finer spatio-temporal scale, pathogen genomic data can provide valuable, often unique insights into disease epidemiology and can be used to infer transmission trees (who-infected-whom), identify super-spreaders, elucidate host contact networks and unravel potential transmission pathways (Kao et al. 2014; Dudas and Bedford 2019). Such information is crucial in improving our understanding of how pathogens spread within and between host populations, which in turn can be used to predict future disease risk, develop effective management interventions and potentially mitigate disease outbreaks. Whole-genome sequencing of pathogens have increasingly been used for epidemiological investigations of human health importance (Ebola virus, Gire et al. 2014; Zika virus, Thézé et al. 2018). Transmission networks in an infectious disease outbreak can now be inferred with more accuracy via identification of genomic variants shared between individuals (Worby et al. 2017) and SNP typing (Stucki et al. 2015).
The emerging field of phylodynamics coupled with parasite genomic data shows great potential for building upon and refining our understanding of parasite transmission in wildlife populations (see earlier section on ‘Spatial structure’). Additionally, incorporating social or contact data with parasite genomic data in a network analysis framework is an exciting open avenue for investigating disease dynamics (White et al. 2017; Gilbertson et al. 2018). For example, in a well-characterized European badger and cattle population, Crispell et al. (2019) compared Mycobacterium bovis (causative agent for bovine tuberculosis) genomes from cattle and badgers to examine cross-species transmission and obtain fine-scale resolution of contact networks for bovine tuberculosis transmission. With pathogen genomic data, researchers were able to quantify the direction and extent of disease transmission, revealing that M. bovis likely transmitted more frequently from badgers to cattle (10.4x in the most likely model) and within-species transmission was more common than between-species disease transmission. However, it is important to note that despite the greater resolution on pathogen transmission events obtained using whole-genome sequencing approaches, it is still challenging to infer fine-scale epidemiological processes, especially for pathogens (e.g. bacteria) that do not evolve fast enough over epidemiological timescales, and might lack informative mutations needed to differentiate them among infected individuals (Kao et al. 2014; Campbell et al. 2018).
Host–parasite interactions: genomewide associations
Pathogens exert a strong selective force on host populations and pathogen mediated selection on host genomes has been widely recognized across a wide range of taxa (Enard et al. 2016; Shultz and Sackton 2019). Recent developments in high-throughput sequencing have allowed researchers to expand beyond a limited number of genes and use whole-genome scans to infer host–parasite interactions. While candidate gene approach targeting immunity related genes (MHCs, TLRs etc., see section on ‘Host genetic diversity’) has been the standard approach to study signatures of parasite-mediated selection on hosts, high-throughput sequencing and genomewide approaches enable an unbiased detection of areas of the genome under selection (e.g., by identifying loci that exhibit outlier allele frequencies).
Whole-genome scans provide a powerful approach as it does not require a priori knowledge about the regions of the genome that are potentially under selection and could lead to identification of novel candidate loci associated with disease susceptibility. For instance, Epstein et al. (2016) identified two genomic regions, with putative cancer or immune function in other mammals, that may be under strong selection imposed by devil facial tumour disease in Tasmanian devils (Sarcophilus harrisii), reflecting a rapid evolutionary response to infection. In another study, Cassin-Sackett et al. (2019) used a hybridization capture approach and SNP typing to identify genes under selection from avian malaria between low-elevation and high-elevation Hawaii amakihi population. The study revealed signatures of selection in immune-related genes and identified several novel candidate loci that may confer tolerance to avian malaria infection in the low-elevation amakihi populations. Overall, for accurate and unbiased estimation of genomic variants associated with disease susceptibility, it is important to account for the underlying population structure and/or environmental heterogeneity. It is also often challenging to assess whether selection on a particular genomic region is parasite-mediated or result of some other environmental factor.
Host–parasite interactions: functional genomics
As discussed above, while genomewide association studies have an enormous potential to reveal novel candidate loci associated with disease susceptibility and/or resistance in natural populations, they are correlational and thus, gene expression studies are still needed to substantiate the underlying mechanisms for variation in host susceptibility to infection. Transcriptomic or RNA-seq approaches have emerged as excellent novel approaches to gain a mechanistic understanding of host–pathogen interactions, particularly for elucidating host response to an infection and/or identifying genes underlying parasite virulence. Host gene expression studies have major implications for elucidating genetic underpinnings of variation in host susceptibility to infection. One can also design effective disease management strategies, by identifying candidate host immune genes/pathways associated with resistance to infection and/or virulence factors expressed by the pathogen, providing potential blocking targets for therapeutics.
RNA-seq allows sequencing of all expressed transcripts in a host organism and examine genes that may be upregulated or downregulated in response to an infection. For instance, Videvall et al. (2015) examined host transcriptome response of European siskins (Carduelis spinus) infected with avian malaria parasites (Plasmodium ashfordi) and identified several genes (e.g., genes related to immune response, stress response) that were differentially expressed at different time-points over the course of infection. In another study examining genome-scale host gene expression patterns underlying amphibian Chytridiomycosis, researchers revealed striking differences in transcriptional response by susceptible and resistant amphibian species exposed to B. dendrobatidis (Poorten and Rosenblum 2016; Eskew et al. 2018). The resistant amphibian host species showed minor changes in gene expression in response to B. dendrobatidis infection but exhibited an upregulation of skin structural integrity pathways compared to the susceptible species.
With further advancements in high-throughput sequencing, dual RNA-seq approaches can now facilitate the simultaneous study of gene expression in hosts and pathogens, and characterize genes underlying resistance/tolerance or identify virulence factors associated with the pathogen (Westermann et al. 2012). Transcriptomic studies generally generate millions of reads from the target species, but a small fraction of reads can also correspond to other nontarget species, including known or unknown pathogens, which can be easily separated by bioinformatics post-processing. For example, in the case of bat WNS, Field et al. (2015) were able to simultaneously characterize transcriptome-wide changes in gene expression in the little brown bats infected with WNS and the causative fungal agent P. destructans.
Concluding remarks
The importance of parasites has long been recognized, and the earliest written records of parasitic infections from ancient Egypt date back to 1500 BC (Cox 2002). However, it has only been over the last few decades that parasites have been recognized as being integral members of ecological systems, playing critical roles in many ecological, evolutionary and ecosystem-level processes. Consequently, there is a growing recognition that disease ecology can contribute substantially to the field of conservation biology. Disease ecology is a fundamentally interdisciplinary field of research and has successfully integrated theoretical models and empirical data from numerous fields, including epidemiology, parasitology, ecology, evolutionary biology and the spatial sciences (e.g., global information systems). Disease ecology has been particularly successful in utilizing the power of molecular ecology to elucidate how ecological and evolutionary factors affect, and are in turn affected by, disease dynamics in natural populations.
Modern genetic and genomic tools improve parasite diagnostics, help better understand parasite transmission and characterize aspects of the parasite (virulence genes) and host (immunogenetics) that affect disease dynamics (DeCandia et al. 2018). Consequently, the integration of disease and molecular ecology has already helped, or has the potential to help, integrate parasites into the broad framework of conservation biology in five major ways. First, at the most basic level, molecular approaches provide an efficient way to unambiguously identify parasites and quantify infection intensities (see above). Second, genetic approaches provide critical information to elucidate coevolutionary dynamics between symbiotic species, such as hosts and parasites. Such information is especially important from a conservation perspective because, in the ongoing mass extinction, parasites and mutualists may comprise some of the most vulnerable organismal groups (Dunn et al. 2009; Derne et al. 2018). Third, molecular tools in conjunction with population genetic and genomic analyses have provided critical insights into how anthropogenic changes, such as global climate change and habitat modification, are likely to affect parasite transmission dynamics (see details above). Additionally, integration of modern technologies (e.g., next-generation sequencing) and growing utilization of analytical approaches (e.g., phylodynamics and network analyses) are allowing conservation biologists to leverage parasite genetics (e.g., bacterial transmission networks) to obtain vital information on host ecology (e.g., social structure and movement patterns) that can be used to inform conservation action (see details above). Fourth, both traditional (e.g., Sanger sequencing) and modern (e.g., next-generation sequencing) technologies provide an excellent platform for further empirical studies of parasites at ecosystem levels, and thus have the potential to improve our understanding of the role parasites play in ecosystem structure and function, and aid further the development of parasites as indicators of ecosystem health (see details above). Finally, conservation biology has long recognized the importance of biodiversity (Soulé 1985) due to its intrinsic value (Ghilarov 2000) and its importance from a utilitarian anthropocentric perspective (e.g., maintenance of ecosystem function and provision of ecosystem services; Justus et al. 2009; Mace et al. 2012). A large proportion of Earth’s biodiversity is likely represented by parasitic species (Poulin and Morand 2000). It is estimated that helminth parasites alone have 50% more species than there are vertebrate hosts (Dobson et al. 2008), but only about 10% of parasite species have been documented (Brooks and Hoberg 2013), and it remains impossible to quantify the exact number of parasitic species. However, molecular tools can be effectively used to uncover cryptic parasite diversity and can thus be used to set conservation priorities through the development of unbiased metrics, such as those based on phylogenetic diversity (Faith 2008; Mooers et al. 2008; Winter et al. 2013; Nunes et al. 2015). Consequently, there remains a great scope for genetic and genomic tools to help incorporate parasites into the broader discussion on species diversity, and potentially help elevate parasites to conservation endpoints in and of themselves.
References
Acevedo-Whitehouse K. and Cunningham A. A. 2006 Is MHC enough for understanding wildlife immunogenetics? Trends Ecol. Evol. 21, 433–438.
Agbali M., Reichard M., Bryjová A., Bryja J. and Smith C. 2010 Mate choice for nonadditive genetic benefits correlate with mhc dissimilarity in the rose bitterling (Rhodeus ocellatus). Evolution 64, 1683–1696.
Altizer S., Nunn C. L., Thrall P. H., Gittleman J. L., Antonovics J., Cunningham A. A. et al. 2003 Social organization and parasite risk in mammals: integrating theory and empirical studies. Annu. Rev. Ecol. Evol. Syst. 34, 517–547.
Amundsen P. A., Lafferty K. D., Knudsen R., Primicerio R., Klemetsen A. and Kuris A. M. 2009 Food web topology and parasites in the pelagic zone of a subarctic lake. J. Anim. Ecol. 78, 563–572.
Andrews K. R., Good J. M., Miller M. R., Luikart G. and Hohenlohe P. A. 2016 Harnessing the power of RADseq for ecological and evolutionary genomics. Nat. Rev. Genet. 17, 81–92.
Archie E. A., Luikart G. and Ezenwa V. O. 2009 Infecting epidemiology with genetics: a new frontier in disease ecology. Trends Ecol. Evol. 24, 21–30.
Atkinson C. T., Dusek R. J., Woods K. L. and Iko W. M. 2000 Pathogenicity of avian malaria in experimentally- infected Hawaii Amakihi. J. Wildl. Dis. 36, 197–204.
Atkinson C. T., Saili K. S., Utzurrum R. B. and Jarvi S. I. 2013 Experimental evidence for evolved tolerance to avian malaria in a wild population of low elevation Hawai’i ’Amakihi (Hemignathus virens). Ecohealth 10, 366–375.
Avramenko R. W., Redman E. M., Lewis R., Yazwinski T. A., Wasmuth J. D. and Gilleard J. S. 2015 Exploring the gastrointestinal “nemabiome”: deep amplicon sequencing to quantify the species composition of parasitic nematode communities. PLoS One 10, 1–18.
Ayres J. S. and Schneider D. S. 2009 The role of anorexia in resistance and tolerance to infections in Drosophila. PLoS Biol. 7, 1–10.
Ayres J. S. and Schneider D. S. 2012 Tolerance of infections. Annu. Rev. Immunol. 30, 271–94.
Babayan S. A., Orton R. J. and Streicker D. G. 2018 Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science 362, 577–580.
Baele G., Suchard M. A., Rambaut A. and Lemey P. 2017 Emerging concepts of data integration in pathogen phylodynamics. Syst. Biol. 66, e47–e65.
Baker K. S., Leggett R. M., Bexfield N. H., Alston M., Daly G., Todd S. et al. 2013 Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology 441, 95–106.
Ballingall K. T., McIntyre A., Lin Z., Timmerman N., Matthysen E., Lurz P. W. W. et al. 2016 Limited diversity associated with duplicated class II MHC-DRB genes in the red squirrel population in the United Kingdom compared with continental Europe. Conserv. Genet. 17, 1171–1182.
Barrow L. N., Allen J. M., Huang X., Bensch S. and Witt C. C. 2018 Genomic sequence capture of haemosporidian parasites: methods and prospects for enhanced study of host-parasite evolution. Mol. Ecol. Resour. 19, 400–441.
Bass D., Stentiford G. D., Littlewood D. T. J. and Hartikainen H. 2015 Diverse applications of environmental DNA methods in parasitology. Trends Parasitol. 31, 499–513.
Bass D., Stentiford G. D., Wang H. C., Koskella B. and Tyler C. R. 2019 The pathobiome in animal and plant diseases. Trends Ecol. Evol. 34, 996–1008.
Bateson Z. W., Hammerly S. C., Johnson J. A., Morrow M. E., Whittingham L. A. and Dunn P. O. 2016 Specific alleles at immune genes, rather than genome-wide heterozygosity, are related to immunity and survival in the critically endangered Attwater’s prairie-chicken. Mol. Ecol. 25, 4730–4744.
Becker M. H., Walke J. B., Cikanek S., Savage A. E., Mattheus N., Santiago C. N. et al. 2015 Composition of symbiotic bacteria predicts survival in Panamanian golden frogs infected with a lethal fungus. Proc. R. Soc. B Biol. Sci. 282.
Bell A. S. and Ranford-Cartwright L. C. 2002 Real-time quantitative PCR in parasitology. Trends Parasitol. 18, 338–342.
Berdoy M., Webster J. P. and Mcdonald D. W. 2000 Fatal attraction in rats infected with Toxoplasma gondii. Proc. R. Soc. B Biol. Sci. 267, 1591–1594.
Bergner L. M., Orton R. J., da Silva Filipe A., Shaw A. E., Becker D. J., Tello C. et al. 2019 Using noninvasive metagenomics to characterize viral communities from wildlife. Mol. Ecol. Resour. 19, 128–143.
Bermejo M., Rodríguez-Teijeiro J. D., Illera G., Barroso A., Vilà C. and Walsh P. D. 2006 Ebola outbreak killed 5000 gorillas. Science 314, 1564.
Biek R. and Real L. A. 2010 The landscape genetics of infectious disease emergence and spread. Mol. Ecol. 19, 3515–3531.
Biek R., Drummond A. J. and Poss M. 2006 A virus reveals population structure and recent demographic history of its carnivore host. Science 311, 538–541.
Biek R., O’hare A., Wright D., Mallon T. and Mccormick C. 2012 Whole genome sequencing reveals local transmission patterns of Mycobacterium bovis in sympatric cattle and badger populations. PLoS Pathog. 8, 1003008.
Bird B. H. and Mazet J. A. K. 2018 Detection of emerging zoonotic pathogens: an integrated one health approach. Annu. Rev. Anim. Biosci. 6, 121–139.
Blackburn T. M. and Ewen J. G. 2017 Parasites as drivers and passengers of human-mediated biological invasions. Ecohealth 14, 61–73.
Blakeslee A. M. H. and Byers J. E. 2008 Using parasites to inform ecological history: comparisons among three congeneric marine snails. Ecology 89, 1068–1078.
Blanar C. A., Munkittrick K. R., Houlahan J., MacLatchy D. L. and Marcogliese D. J. 2009 Pollution and parasitism in aquatic animals: a meta-analysis of effect size. Aquat. Toxicol. 93, 18–28.
Blanchong J. A., Scribner K. T., Kravchenko A. N. and Winterstein S. R. 2007 TB-infected deer are more closely related than non-infected deer. Biol. Lett. 3, 104–106.
Blaustein A. R., Gervasi S. S., Johnson P. T. J., Hoverman J. T., Belden L. K., Bradley P. W. and Xie G. Y. 2012 Ecophysiology meets conservation: understanding the role of disease in amphibian population declines. Philos. Trans. R. Soc. B Biol. Sci. 367, 1688–1707.
Blehert D. S., Hicks A. C., Behr M., Meteyer C. U., Berlowski-Zier B. M., Buckles E. L. et al. 2009 Bat white-nose syndrome: An emerging fungal pathogen? Science 323, 227.
Bletz M. C., Loudon A. H., Becker M. H., Bell S. C., Woodhams D. C., Minbiole K. P. C. et al. 2013 Mitigating amphibian chytridiomycosis with bioaugmentation: characteristics of effective probiotics and strategies for their selection and use. Ecol. Lett. 16, 807–820.
Bodewes R., Ruiz-Gonzalez A., Schapendonk C. M. E., Van Den Brand J. M. A., Osterhaus A. D. M. E. et al. 2014 Viral metagenomic analysis of feces of wild small carnivores. Virol. J. 11, 1–13.
Bohmann K., Schnell I. B. and Gilbert M. T. P. 2013 When bugs reveal biodiversity. Mol. Ecol. 22, 909–911.
Bohmann K., Evans A., Gilbert M. T. P., Carvalho G. R., Creer S., Knapp M. et al. 2014 Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 29, 358–367.
Boots M. and Sasaki A. 2003 Parasite evolution and extinctions. Ecol. Lett. 6, 176–182.
Bouyer J., Dicko A. H., Cecchi G., Ravel S., Guerrini L., Solano P. et al. 2015 Mapping landscape friction to locate isolated tsetse populations that are candidates for elimination. Proc. Natl. Acad. Sci. USA 112, 14575–14580.
Brambilla A., Keller L., Bassano B. and Grossen C. 2018 Heterozygosity–fitness correlation at the major histocompatibility complex despite low variation in Alpine ibex (Capra ibex). Evol. Appl. 11, 631–644.
Brandt A. L., Green M. L., Ishida Y., Roca A. L., Novakofski J. and Mateus-Pinilla N. E. 2018 Influence of the geographic distribution of prion protein gene sequence variation on patterns of chronic wasting disease spread in white-tailed deer (Odocoileus virginianus). Prion 12, 204–215.
Brooks D. R. and Hoberg E. P. 2013 The emerging infectious diseases crisis and pathogen pollution. Balanc. Nat. Hum. Impact 215–229.
Brown B. L., Creed R. P., Skelton J., Rollins M. A. and Farrell K. J. 2012 The fine line between mutualism and parasitism: complex effects in a cleaning symbiosis demonstrated by multiple field experiments. Oecologia 170, 199–207.
Brunker K., Lemey P., Marston D. A., Fooks A. R., Lugelo A., Ngeleja C. et al. 2018 Landscape attributes governing local transmission of an endemic zoonosis: rabies virus in domestic dogs. Mol. Ecol. 27, 773–788.
Buck J. C. and Ripple W. J. 2017 Infectious agents trigger trophic cascades. Trends Ecol. Evol. 32, 681–694.
Bull C. M., Godfrey S. S. and Gordon D. M. 2012 Social networks and the spread of Salmonella in a sleepy lizard population. Mol. Ecol. 21, 4386–4392.
Cable J., Barber I., Boag B., Ellison A. R., Morgan E. R., Murray K. et al. 2017 Global change, parasite transmission and disease control: Lessons from ecology. Philos. Trans. R. Soc. B Biol. Sci. 372.
Caillaud D., Craft M. E. and Meyers L. A. 2013 Epidemiological effects of group size variation in social species. J. R. Soc. Interface 10.
Caillaud D., Levréro F., Cristescu R., Gatti S., Dewas M., Douadi M. et al. 2006 Gorilla susceptibility to Ebola virus: the cost of sociality. Curr. Biol. 16, 489–491.
Caldwell A. and Siddle H. V. 2017 The role of MHC genes in contagious cancer: the story of Tasmanian devils. Immunogenetics 69, 537–545.
Caldwell R. M., Schafer J. F., Compton L. E. and Patterson F. L. 1958 Tolerance to cereal leaf rusts. Science 128, 714–715.
Calegaro-Marques C. and Amato S. B. 2014 Urbanization breaks up host-parasite interactions: A case study on parasite community ecology of rufous-bellied thrushes (Turdus rufiventris) along a rural-urban gradient. PLoS One 9.
Campbell F., Strang C., Ferguson N., Cori A. and Jombart T. 2018 When are pathogen genome sequences informative of transmission events? PLoS Pathog. 14.
Carpi G., Cagnacci F., Wittekindt N. E., Zhao F., Qi J., Tomsho L. P. et al. 2011 Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS One 6.
Cassin-Sackett L., Callicrate T. E. and Fleischer R. C. 2019 Parallel evolution of gene classes, but not genes: Evidence from Hawai’ian honeycreeper populations exposed to avian malaria. Mol. Ecol. 28, 568–583.
Chen W., Bei Y. and Li H. 2015 Genetic variation of the major histocompatibility complex (MHC class II B gene) in the threatened Hume’s pheasant, Syrmaticus humiae. PLoS One 10, 1–12.
Chernin E. 1968 Interference with the capacity of Schistosoma mansoni Miracidia to infect the molluscan host. J. Parasitol. 54, 509.
Chiu L., Bazin T., Truchetet M. E., Schaeverbeke T., Delhaes L. and Pradeu T. 2017 Protective microbiota: From localized to long-reaching co-immunity. Front. Immunol. 8, 1678.
Chiyo P. I., Grieneisen L. E., Wittemyer G., Moss C. J., Lee P. C., Douglas-Hamilton I. et al. 2014 The influence of social structure, habitat, and host traits on the transmission of Escherichia coli in wild elephants. PLoS One 9.
Civitello D. J., Cohen J., Fatima H., Halstead N. T., Liriano J., McMahon T. A. et al. 2015 Biodiversity inhibits parasites: Broad evidence for the dilution effect. Proc. Natl. Acad. Sci. USA 112, 8667–8671.
Clark N. J., Clegg S. M. and Lima M. R. 2014 A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): new insights from molecular data. Int. J. Parasitol. 44, 329–338.
Cleaveland S., Packer C., Hampson K., Kaare M., Kock R., Craft M. et al. 2009 The multiple roles of infectious diseases in the serengeti ecosystem. In Serengeti III: human impacts on ecosystem dynamics (ed. A. R. E. Sinclair, C. Packer, S. A. R. Mduma and J. M. Fryxell), pp. 209–241. University of Chicago Press, Chicago.
Coates A., Barnett L. K., Hoskin C. and Phillips B. L. 2017 Living on the edge: parasite prevalence changes dramatically across a range edge in an invasive gecko. Am. Nat. 189, 178–183.
Cobb R. C., Chan M. N., Meentemeyer R. K. and Rizzo D. M. 2012 Common factors drive disease and coarse woody debris dynamics in forests impacted by sudden oak death. Ecosystems 15, 242–255.
Connelly S., Pringle C. M., Bixby R. J., Brenes R., Whiles M. R., Lips K. R. et al. 2008 Changes in stream primary producer communities resulting from large-scale catastrophic amphibian declines: Can small-scale experiments predict effects of tadpole loss? Ecosystems 11, 1262–1276.
Côté S. D., Stien A., Irvine R. J., Dallas J. F., Marshall F., Halvorsen O. et al. 2005 Resistance to abomasal nematodes and individual genetic variability in reindeer. Mol. Ecol. 14, 4159–4168.
Crabtree M. B., Kading R. C., Mutebi J.-P., Lutwama J. J. and Miller B. R. 2013 Identification of host blood from engorged mosquitoes collected in western uganda using cytochrome oxidase I gene sequences. J. Wildl. Dis. 49, 611–626.
Craft M. E. 2015 Infectious disease transmission and contact networks in wildlife and livestock. Philos. Trans. R. Soc. B Biol. Sci. 370.
Crispell J., Benton C. H., Balaz D., De Maio N., Ahkmetova A., Allen A. et al. 2019 Combining genomics and epidemiology to analyse bi-directional transmission of Mycobacterium bovis in a multi-host system. Elife 8, 1–36.
Cullingham C. I., Merrill E. H., Pybus M. J., Bollinger T. K., Wilson G. A. and Coltman D. W. 2011a Broad and fine-scale genetic analysis of white-tailed deer populations: estimating the relative risk of chronic wasting disease spread. Evol. Appl. 4, 116–131.
Cullingham C. I., Nakada S. M., Merrill E. H., Bollinger T. K., Pybus M. J. and Coltman D. W. 2011b Multiscale population genetic analysis of mule deer (Odocoileus hemionus hemionus) in western Canada sheds new light on the spread of chronic wasting disease. Can. J. Zool. 89, 134–147.
Daszak P., Cunningham A. and Hyatt A. 2000 Emerging infectious diseases of wildlife - threats to biodiversity and human health. Science 287, 443–449.
De Castro F. and Bolker B. 2005 Mechanisms of disease-induced extinction. Ecol. Lett. 8, 117–126.
De Roode J. C. and Lefèvre T. 2012 Behavioral immunity in insects. Insects 3, 789–820.
Decaestecker E., Gaba S., Raeymaekers J. A. M., Stoks R., Van Kerckhoven L., Ebert D. et al. 2007 Host-parasite “Red Queen” dynamics archived in pond sediment. Nature 450, 870–873.
DeCandia A. L., Dobson A. P. and VonHoldt B. M. 2018 Toward an integrative molecular approach to wildlife disease. Conserv. Biol. 32, 798–807.
Dellicour S., Rose R. and Pybus O. G. 2016 Explaining the geographic spread of emerging epidemics: A framework for comparing viral phylogenies and environmental landscape data. BMC Bioinformatics 17, 1–12.
Derne B. T., Hutchinson M. N., Weinstein P., Gardner M. G. and Halliday B. 2018 Parasite in peril? A new species of mite in the genus Ophiomegistus Banks (Parasitiformes: Paramegistidae) on an endangered host, the pygmy bluetongue lizard Tiliqua adelaidensis (Peters) (Squamata: Scincidae). Austral Ecol. 44, 420–432.
Deyoung R. W., Zamorano A., Mesenbrink B. T., Campbell T. A., Leland B. R., Moore G. M. et al. 2009 Landscape-genetic analysis of population structure in the texas gray fox oral rabies vaccination zone. J. Wildl. Manage. 73, 1292–1299.
Dharmarajan G. 2015 Inbreeding in stochastic subdivided mating systems: the genetic consequences of host spatial structure, aggregated transmission dynamics and life history characteristics in parasite populations. J. Genet. 94, 43–53.
Dharmarajan G., Beasley J. C. and Rhodes O. E. 2010 Spatial and temporal factors affecting parasite genotypes encountered by hosts: empirical data from American dog ticks (Dermacentor variabilis) parasitising raccoons (Procyon lotor). Int. J. Parasitol. 40, 787–795.
Dharmarajan G., Beasley J. C. and Rhodes O. E. 2011 Heterozygote deficiencies in parasite populations: an evaluation of interrelated hypotheses in the raccoon tick, Ixodes texanus. Heredity 106, 253–260.
Dharmarajan G., Beasley J. C., Fike J. A., Raizman E. A., Wu C. C., Pogranichniy R. M. et al. 2012 Effects of kin-structure on disease dynamics in raccoons (Procyon lotor) inhabiting a fragmented landscape. Basic Appl. Ecol. 13, 560–567.
Dharmarajan G., Beasley J. C., Beatty W. S., Olson Z. H., Fike J. A., Rhodes O. E. et al. 2016 Genetic co-structuring in host-parasite systems: Empirical data from raccoons and raccoon ticks. Ecosphere 7, 1–15.
Dharmarajan G., Walker K. D. and Lehmann T. 2019 Variation in tolerance to parasites affects vectorial capacity of natural asian tiger mosquito populations. Curr. Biol. 29, 3946–3952.
Dheilly N. M., Bolnick D., Bordenstein S., Brindley P. J., Figuères C., Holmes E. C. et al. 2017 Parasite microbiome project: systematic investigation of microbiome dynamics within and across parasite-host interactions. MSystems 2, e00050-17.
Dheilly N. M., Martínez Martínez J., Rosario K., Brindley P. J., Fichorova R. N., Kaye J. Z. et al. 2019 Parasite microbiome project: Grand challenges. PLoS Pathog. 15, e1008028.
Dhondt A. A., Altizer S., Cooch E. G., Davis A. K., Dobson A. and Driscoll M. J. L. 2005 Dynamics of a novel pathogen in an avian host: Mycoplasmal conjunctivitis in house finches. Acta Trop. 94, 77–93.
Dirzo R., Young H. S., Galetti M., Ceballos G., Isaac N. J. B. and Collen B. 2014 Defaunation in the Anthropocene. Science 345, 401–406.
Dobson A. and Crawley M. 1994 Pathogens and the structure of plant communities. Trends Ecol. Evol. 9, 393–398.
Dobson A., Lafferty K. D., Kuris A. M., Hechinger R. F. and Jetz W. 2008 Homage to Linnaeus: How many parasites? How many hosts? Proc. Natl. Acad. Sci. USA 105, 11482–11489.
Dorigatti I., McCormack C., Nedjati-Gilani G. and Ferguson N. M. 2018 Using Wolbachia for Dengue Control: Insights from Modelling. Trends Parasitol. 34, 102–113.
Drees K. P., Lorch J. M. Puechmaille S. J., Parise K. L., Wibbelt G., Hoyt J. R. et al. 2017 Phylogenetics of a fungal invasion: origins and widespread dispersal of white-nose syndrome. MBio 8, e01941-17.
Dudas G. and Bedford T. 2019 The ability of single genes vs full genomes to resolve time and space in outbreak analysis. BMC Evol. Biol. 19, 1–17.
Dunn A. M., Torchin M. E., Hatcher M. J., Kotanen P. M., Blumenthal D. M., Byers J. E. et al. 2012 Indirect effects of parasites in invasions. Funct. Ecol. 26, 1262–1274.
Dunn R. R., Harris N. C., Colwell R. K., Koh L. P. and Sodhi N. S. 2009 The sixth mass coextinction: Are most endangered species parasites and mutualists? Proc. R. Soc. B Biol. Sci. 276, 3037–3045.
Duvallet C., Gibbons S. M., Gurry T., Irizarry R. A. and Alm E. J. 2017 Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 8, 1–10.
Eastwood J. R., Ribot R. F. H., Rollins L. A., Buchanan K. L., Walder K., Bennett A. T. D. et al. 2017 Host heterozygosity and genotype rarity affect viral dynamics in an avian subspecies complex. Sci. Rep. 7, 1–11.
Elbers J. P. and Taylor S. S. 2016 Major histocompatibility complex polymorphism in reptile conservation. Herpetol. Conserv. Biol. 11, 1–12.
Elton C. S. 1958 The Ecology of Invasions by Animals and Plants, The Ecology of Invasions by Animals and Plants, pp. 1–159. Springer, Boston, MA.
Enard D., Cai L., Gwennap C. and Petrov D. A. 2016 Viruses are a dominant driver of protein adaptation in mammals. Elife 5, e12469.
Epstein B., Jones M., Hamede R., Hendricks S., McCallum H., Murchison E. P. et al. 2016 Rapid evolutionary response to a transmissible cancer in Tasmanian devils. Nat. Commun. 7, 12684.
Eskew E. A., Shock B. C., Ladouceur E. E. B., Keel K., Miller M. R., Foley J. E. et al. 2018 Gene expression differs in susceptible and resistant amphibians exposed to Batrachochytrium dendrobatidis. R. Soc. Open Sci. 5, 170910.
Eszterbauer E., Forró B., Tolnai Z., Guti C. F., Zsigmond G., Hoitsy G. and Kallert D. M. 2015 Parental genetic diversity of brown trout (Salmo trutta m. fario) brood stock affects offspring susceptibility to whirling disease. Parasit. Vectors 8, 1–9.
Ewald P. W. 1987 Transmission modes and evolution of the parasitism-mutualism continuum. Ann. N. Y. Acad. Sci. 503, 295–306.
Ewen J. G., Bensch S., Blackburn T. M., Bonneaud C., Brown R., Cassey P. et al. 2012 Establishment of exotic parasites: The origins and characteristics of an avian malaria community in an isolated island avifauna. Ecol. Lett. 15, 1112–1119.
Faith D. P. 2008 Threatened species and the potential loss of phylogenetic diversity: conservation scenarios based on estimated extinction probabilities and phylogenetic risk analysis. Conserv. Biol. 22, 1461–1470.
Faria N. R., Suchard M. A., Rambaut A. and Lemey P. 2011 Toward a quantitative understanding of viral phylogeography. Curr. Opin. Virol. 1, 423–429.
Field K. A., Johnson J. S., Lilley T. M., Reeder S. M., Rogers E. J., Behr M. J. et al. 2015 The white-nose syndrome transcriptome: activation of anti-fungal host responses in wing tissue of hibernating little brown myotis. PLoS Pathog. 11, 1–29.
Fisher M. C., Henk D. A., Briggs C. J., Brownstein J. S., Madoff L. C., McCraw S. L. et al. 2012 Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 1–18.
Flaherty B. R., Talundzic E., Barratt J., Kines K. J., Olsen C., Lane M. et al. 2018 Restriction enzyme digestion of host DNA enhances universal detection of parasitic pathogens in blood via targeted amplicon deep sequencing. Microbiome 6, 164.
Frank S. A. 1996 Models of parasite virulence. Q. Rev. Biol. 71, 37–78.
Frick W. F., Pollock J. F., Hicks A. C., Langwig K. E., Reynolds D. S., Turner G. G. et al. 2010 An emerging disease causes regional population collapse of a common north american bat species. Science 329, 679–682.
Fu M. and Waldman B. 2017 Major histocompatibility complex variation and the evolution of resistance to amphibian chytridiomycosis. Immunogenetics 69, 529–536.
Gariepy T. D., Lindsay R., Ogden N. and Gregory T. R. 2012 Identifying the last supper: Utility of the DNA barcode library for bloodmeal identification in ticks. Mol. Ecol. Resour. 12, 646–652.
Ghai R. R., Fugère V., Chapman C. A., Goldberg T. L. and Davies T. J. 2015 Sickness behaviour associated with non-lethal infections in wild primates. Proc. R. Soc. B Biol. Sci. 282.
Ghilarov A. M. 2000 Ecosystem functioning and intrinsic value of biodiversity. Oikos 90, 408–412.
Gilbert J. A., Jansson J. K. and Knight R. 2014 The Earth Microbiome project: successes and aspirations. BMC Biol. 12, 1–4.
Gilbertson M. L. J., Fountain-Jones N. M. and Craft M. E. 2018 Incorporating genomic methods into contact networks to reveal new insights into animal behaviour and infectious disease dynamics. Behaviour 155, 759–791.
Gire S. K., Goba A., Andersen K. G., Sealfon R. S. G., Park D. J., Kanneh L. et al. 2014 Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345, 1369–1372.
Gopinath S., Lichtman J. S., Bouley D. M., Elias J. E. and Monack D. M. 2014 Role of disease-associated tolerance in infectious superspreaders. Proc. Natl. Acad. Sci. USA 111, 15780–15785.
Grenfell B. T., Pybus O. G., Gog J. R., Wood J. L. N., Daly J. M., Mumford J. A. et al 2004 Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303, 327–333.
Griffin R. H. and Nunn C. L. 2012 Community structure and the spread of infectious disease in primate social networks. Evol. Ecol. 26, 779–800.
Hamer G. L., Kitron U. D., Goldberg T. L., Brawn J. D., Loss S. R. et al. 2009 Host selection by Culex pipiens mosquitoes and west nile virus amplification. Am. J. Trop. Med. Hyg. 80, 268–278.
Harris R. N., Brucker R. M., Walke J. B., Becker M. H., Schwantes C. R., Flaherty D. C. et al. 2009 Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 3, 818–824.
Hart B. L. 1990 Behavioral adaptations to pathogens and parasites: five strategies. Neurosci. Biobehav. Rev. 14, 273–294.
Hatcher M. J., Dick J. T. A. and Dunn A. M. 2012a Disease emergence and invasions. Funct. Ecol. 26, 1275–1287.
Hatcher M. J., Dick J. T. and Dunn A. M. 2012b Diverse effects of parasites in ecosystems: linking interdependent processes. Front. Ecol. Environ. 10, 186–194.
Hatcher M. J., Dick J. T. A. and Dunn A. M. 2014 Parasites that change predator or prey behaviour can have keystone effects on community composition. Biol. Lett. 10, 1–4.
Hawley D. M. and Altizer S. M. 2011 Disease ecology meets ecological immunology: Understanding the links between organismal immunity and infection dynamics in natural populations. Funct. Ecol. 25, 48–60.
Hedrick P. W., Kim T. J. and Parker K. M. 2001 Parasite resistance and genetic variation in the endangered Gila topminnow. Anim. Conserv. 4, 103–109.
Hellgren O., Bensch S. and Malmqvist B. 2008 Bird hosts, blood parasites and their vectors - Associations uncovered by molecular analyses of blackfly blood meals. Mol. Ecol. 17, 1605–1613.
Heloise C., Garant D., Robert K., Mainguy J. and Pelletier F. 2012 Genetic structure and rabies spread potential in raccoons: The role of landscape barriers and sex-biased dispersal. Evol. Appl. 5, 393–404.
Hess A. D. and Hayes R. O. 1970 Relative potentials of domestic animals for zooprophylaxis against mosquito vectors of encephalitis. Am. J. Trop. Med. Hyg. 19, 327–334.
Hilker F. M., Lewis M. A., Seno H., Langlais M. and Malchow H. 2005 Pathogens can slow down or reverse invasion fronts of their hosts. Biol. Invasions 7, 817–832.
Hill N. J., Takekawa J. Y., Ackerman J. T., Hobson K. A., Herring G., Cardona C. J. et al. 2012 Migration strategy affects avian influenza dynamics in mallards (Anas platyrhynchos). Mol. Ecol. 21, 5986–5999.
Hindson B. J., Ness K. D., Masquelier D. A., Belgrader P., Heredia N. J. and Makarewicz A. J. 2011 High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83, 8604–8610.
Hoberg E. P. and Brooks D. R. 2015 Evolution in action: climate change, biodiversity dynamics and emerging infectious disease. Philos. Trans. R. Soc. B Biol. Sci. 370, 20130553.
Holt E. A. and Miller S. W. 2011 Bioindicators: using organisms to measure environmental impacts. Nat. Educ. Knowl. 3, 8.
Hooper L. V., Littman D. R. and Macpherson A. J. 2012 Interactions between the microbiota and the immune system. Science 336, 1268–1273.
Horwitz P. and Wilcox B. A. 2005 Parasites, ecosystems and sustainability: An ecological and complex systems perspective. Int. J. Parasitol. 35, 725–732.
Hoyt J. R., Cheng T. L., Langwig K. E., Hee M. M., Frick W. F. and Kilpatrick A. M. 2015 Bacteria isolated from bats inhibit the growth of Pseudogymnoascus destructans, the causative agent of white-nose syndrome. PLoS One 10, e0121329.
Hoyt J. R., Langwig K. E., White J. P., Kaarakka H. M., Redell J. A., Parise K. L. et al. 2019 Field trial of a probiotic bacteria to protect bats from white-nose syndrome. Sci. Rep. 9, 1–9.
Hudson P. J., Dobson A. P. and Lafferty K. D. 2006 Is a healthy ecosystem one that is rich in parasites? Trends Ecol. Evol. 21, 381–385.
Huggins L. G., Koehler A. V., Ng-Nguyen D., Wilcox S., Schunack B., Inpankaew T. et al. 2019 A novel metabarcoding diagnostic tool to explore protozoan haemoparasite diversity in mammals: a proof-of-concept study using canines from the tropics. Sci. Rep. 9, 1–10.
Hughes A. L. 1991 Society for conservation biology mhc polymorphism and the design of captive breeding programs. Conserv. Biol. 5, 249–251.
Hughes D. P., Brodeur J. and Thomas F. (eds.) 2012. Host manipulation by parasites. Oxford University Press, Oxford.
Hupalo D. N., Bradic M. and Carlton J. M. 2015 The impact of genomics on population genetics of parasitic diseases. Curr. Opin. Microbiol. 23, 49–54.
Huspeni T. C. and Lafferty K. D. 2004 Using larval trematodes that parasitize snails to evaluate a saltmarsh restoration project. Ecol. Appl. 14, 795–804.
Huver J. R., Koprivnikar J., Johnson P. T. J. and Whyard S. 2015 Development and application of an eDNA method to detect and quantify a pathogenic parasite in aquatic ecosystems. Ecol. Appl. 25, 991–1002.
Jiang T., Shwab E. K., Martin R. M., Gerhold R. W., Rosenthal B. M., Dubey J. P. et al. 2018 A partition of Toxoplasma gondii genotypes across spatial gradients and among host species, and decreased parasite diversity towards areas of human settlement in North America. Int. J. Parasitol. 48, 611–619.
Johnson P. T. J., de Roode J. C. and Fenton A. 2015a Why infectious disease research needs community ecology. Science 349, 1259504.
Johnson P. T. J., Ostfeld R. S. and Keesing F. 2015b Frontiers in research on biodiversity and disease. Ecol. Lett. 18, 1119–1133.
Johnson P. T. J., Dobson A., Lafferty K. D., Marcogliese D. J., Memmott J., Orlofske S. A. et al. 2010 When parasites become prey: ecological and epidemiological significance of eating parasites. Trends Ecol. Evol. 25, 362–371.
Jones K. E., Patel N. G., Levy M. A., Storeygard A., Balk D., Gittleman J. L. and Daszak P. 2008 Global trends in emerging infectious diseases. Nature 451, 990–993.
Jones M. R. and Good J. M. 2016 Targeted capture in evolutionary and ecological genomics. Mol. Ecol. 25, 185–202.
Justus J., Colyvan M., Regan H. and Maguire L. 2009 Buying into conservation: intrinsic versus instrumental value. Trends Ecol. Evol. 24, 187–191.
Kamath P. L., Foster J. T., Drees K. P., Luikart G., Quance C., Anderson N. J. et al. 2016 Genomics reveals historic and contemporary transmission dynamics of a bacterial disease among wildlife and livestock. Nat. Commun. 7, 1–10.
Kamiya T., O’Dwyer K., Westerdahl H., Senior A. and Nakagawa S. 2014 A quantitative review of MHC-based mating preference: The role of diversity and dissimilarity. Mol. Ecol. 23, 5151–5163.
Kamoroff C. and Goldberg C. S. 2017 Using environmental DNA for early detection of amphibian chytrid fungus Batrachochytrium dendrobatidis prior to a ranid die-off. Dis. Aquat. Organ. 127, 75–79.
Kao R. R., Haydon D. T., Lycett S. J. and Murcia P. R. 2014 Supersize me: How whole-genome sequencing and big data are transforming epidemiology. Trends Microbiol. 22, 282–291.
Kauffman E. B., Jones S. A., Dupuis A. P., Ngo K. A., Bernard K. A. and Kramer L. D. 2003 Virus detection protocols for West Nile virus in vertebrate and mosquito specimens. J. Clin. Microbiol. 41, 3661–3667.
Keane R. M. and Crawley M. J. 2002 Exotic plant invasions and the enemy release hypothesis. Trends Ecol. Evol. 17, 164–170.
Keesing F. and Ostfeld R. S. 2012 Disease ecology. In Integrating ecology and poverty reduction (ed. J. C. Ingram, F. DeClerck and C. R. del Rio), pp. 217–230. Springer-Verlag, New York.
Keesing F. and Young T. P. 2014 Cascading consequences of the loss of large mammals in an African Savanna. Bioscience 64, 487–495.
Keith D. A., Rodríguez J. P., Brooks T. M., Burgman M. A., Barrow E. G., Bland L. et al. 2015 The IUCN red list of ecosystems: Motivations, challenges, and applications. Conserv. Lett. 8, 214–226.
Kelly A. C., Mateus-Pinilla N. E., Brown W., Ruiz M. O., Douglas M. R., Douglas M. E. et al. 2014 Genetic assessment of environmental features that influence deer dispersal: Implications for prion-infected populations. Popul. Ecol. 56, 327–340.
Kelly D. W., Paterson R. A., Toensend C. R., Poulin R. and Tompkins D. M. 2009 Parasite spillback: A neglected concept in invasion ecology? Ecology 90, 2047–2056.
King J. L., Smith A. D., Mitchell E. A. and Allen M. S. 2017 Validation of droplet digital PCR for the detection and absolute quantification of Borrelia DNA in Ixodes scapularis ticks. Parasitology 144, 359–367.
King K. C. and Lively C. M. 2012 Does genetic diversity limit disease spread in natural host populations. Heredity 109, 199–203.
Kocher A., Gantier J.-C., Gaborit P., Zinger L., Holota H., Valiere S. et al. 2017 Vector soup: high-throughput identification of Neotropical phlebotomine sand flies using metabarcoding. Mol. Ecol. Resour. 17, 172–182.
Kock R. A., Wambua J. M., Mwanzia J., Wamwayi H., Ndungu E. K., Barrett T. et al. 1999 Rinderpest epidemic in wild ruminants in Kenya 1993–97. Vet. Rec. 145, 275–283.
Koepfli C., Nguitragool W., Hofmann N. E., Robinson L. J., Ome-Kaius M., Sattabongkot J. et al. 2016 Sensitive and accurate quantification of human malaria parasites using droplet digital PCR (ddPCR). Sci. Rep. 6, 39183.
Kozakiewicz C. P., Burridge C. P., Funk W. C., VandeWoude S., Craft M. E., Crooks K. R. et al. 2018 Pathogens in space: Advancing understanding of pathogen dynamics and disease ecology through landscape genetics. Evol. Appl. 11, 1763–1778.
Kuris A. M., Hechinger R. F., Shaw J. C., Whitney K. L., Aguirre-Macedo L., Boch C. A. et al. 2008 Ecosystem energetic implications of parasite and free-living biomass in three estuaries. Nature 454, 515–518.
Kutzer M. A. M. and Armitage S. A. O. 2016 Maximising fitness in the face of parasites: a review of host tolerance. Zoology 119, 281–289.
LaDeau S. L., Kilpatrick A. M. and Marra P. P. 2007 West Nile virus emergence and large-scale declines of North American bird populations. Nature 447, 710–713.
Ladle R. J. 1992 Parasites and Sex: Catching the Red Queen. Trends Ecol. Evol. 7, 405–408.
Lafferty K. D. 1997 Environmental parasitology: what can parasites tell us about human impacts on the environment? Parasitol. Today 13, 251–255.
Lafferty K. D. and Shaw J. C. 2013 Comparing mechanisms of host manipulation across host and parasite taxa. J. Exp. Biol. 216, 56–66.
Lafferty K. D., Dobson A. P. and Kuris A. M. 2006 Parasites dominate food web links. Proc. Natl. Acad. Sci. USA 103, 11211–11216.
Lazzaro B. P. and Clark A. G. 2012 Rapid evolution of innate immune response genes. In Rapidly Evolving Genes and Genetic Systems (ed. R. S. Singh, J. Xu and R. J. Kulathinal), pp 312. Oxford University Press (e-book), Oxford.
le Roex N., Koets A. P., van Helden P. D. and Hoal E. G. 2013 Gene polymorphisms in African buffalo associated with susceptibility to Bovine Tuberculosis infection. PLoS One 8, 1–6.
Lee J. S., Mackie R. S., Harrison T., Shariat B., Kind T., Kehl T. et al. 2017 Targeted Enrichment for Pathogen Detection and Characterization in Three Felid Species. J. Clin. Microbiol. 55, 1658–1670.
Lee P. S., Sing K. W. and Wilson J. J. 2015 Reading mammal diversity from flies: The persistence period of amplifiable mammal mtDNA in blowfly guts (Chrysomya megacephala) and a new DNA mini-barcode target. PLoS One 10, 1–12.
Lefèvre T., Lebarbenchon C., Gauthier-Clerc M., Missé D., Poulin R. and Thomas F. 2009 The ecological significance of manipulative parasites. Trends Ecol. Evol. 24, 41–48.
Leung T. L. F. and Poulin R. 2008 Parasitism, commensalism, and mutualism: exploring the many shades of symbioses. Vie Milieu 58, 107–115.
Leventhal G. E., Kouyos R., Stadler T., von Wyl V., Yerly S., Böni J. et al. 2012 Inferring epidemic contact structure from phylogenetic trees. PLoS Comput. Biol. 8.
Li H., Bai R., Zhao Z., Tao L., Ma M., Ji Z. et al. 2018 Application of droplet digital PCR to detect the pathogens of infectious diseases. Biosci. Rep. 38.
Lindström E. R., Andrén H., Angelstam P., Cederlund G., Jäderberg L. and Lemnell P. 1994 Disease Reveals the predator: sarcoptic mange, red fox predation, and prey populations. Ecology 75, 1042–1049.
Lipkin W. I. 2013 The changing face of pathogen discovery and surveillance. Nat. Rev. Microbiol. 11, 133–141.
Lively C. M. and Dybdahl M. F. 2000 Parasite adaptation to locally common host genotypes. Nature 405, 679–681.
Lively C. M., Craddock C. and Vrijenhoek R. C. 1990 Red Queen hypothesis supported by parasitism in sexual and clonal fish. Nature 344, 864–866.
Lorch J. M., Gargas A., Meteyer C. U., Berlowski-Zier B. M., Green D. E. and Shearn-Bochsler V. 2010 Rapid polymerase chain reaction diagnosis of white-nose syndrome in bats. J. Vet. Diagnostic Investig. 22, 224–230.
Lovett G. M., Arthur M. A., Weathers K. C. and Griffin J. M. 2010 Long-term changes in forest carbon and nitrogen cycling caused by an introduced pest/pathogen complex. Ecosystems 13, 1188–1200.
Luquet E., Garner T. W. J., Léna J. P., Bruel C., Joly P. and Lengagne T. 2012 Genetic erosion in wild populations makes resistance to a pathogen more costly. Evolution 66, 1942–1952.
Lymbery A. J., Morine M., Kanani H. G., Beatty S. J. and Morgan D. L. 2014 Co-invaders: The effects of alien parasites on native hosts. Int. J. Parasitol. Parasites Wildl. 3, 171–177.
Ma M., Huang Y., Gong Z., Zhuang L., Li C., Yang H. et al. 2011 Discovery of DNA viruses in wild-caught mosquitoes using small RNA high throughput sequencing. PLoS One 6, 1–7.
MacDonald D. W. 1979 The flexible social system of the golden jackal, Canis aureus. Behav. Ecol. Sociobiol. 5, 17–38.
Mace G. M., Norris K. and Fitter A. H. 2012 Biodiversity and ecosystem services: a multilayered relationship. Trends Ecol. Evol. 27, 19–25.
Magle S. B., Samuel M. D., Van Deelen T. R., Robinson S. J. and Mathews N. E. 2013 Evaluating spatial overlap and relatedness of white-tailed deer in a chronic wasting disease management zone. PLoS One 8, e56568.
Mamanova L., Coffey A. J., Scott C. E., Kozarewa I., Turner E. H., Kumar A. et al. 2010 Target-enrichment strategies for next-generation sequencing. Nat. Methods 7, 111–118.
Mancini M. V., Damiani C., Accoti A., Tallarita M., Nunzi E., Cappelli A. 2018 Estimating bacteria diversity in different organs of nine species of mosquito by next generation sequencing. BMC Microbiol. 18, 1–10.
Manel S. and Holderegger R. 2013 Ten years of landscape genetics. Trends Ecol. Evol. 28, 614–621.
Marcogliese D. J. 2005 Parasites of the superorganism: are they indicators of ecosystem health? Int. J. Parasitol. 35, 705–716.
Martel A., Spitzen-van der Sluijs A., Blooi M., Bert W., Ducatelle R., Fisher M. C. et al. 2013 Batrachochytrium salamandrivorans sp. nov. causes lethal chytridiomycosis in amphibians. Proc. Natl. Acad. Sci. USA 110, 15325–15329.
Mauricio R., Rausher M. D. and Burdick D. S. 1997 Variation in the defense strategies of plants: Are resistance and tolerance mutually exclusive? Ecology 78, 1301–1311.
McGrew A. K., O’Hara T. M., Stricker C. A., Salman M. D., Van Bonn W., Gulland F. M. D. et al. 2018 Ecotoxicoparasitology of the gastrointestinal tracts of pinnipeds: the effect of parasites on the potential bioavailability of total mercury (THg). Sci. Total Environ. 631–632, 233–238.
Medzhitov R., Schneider D. S. and Soares M. P. 2012 Disease tolerance as a defense strategy. Science 335, 936–941.
Miaud C., Arnal V., Poulain M., Valentini A. and Dejean T. 2019 eDNA increases the detectability of ranavirus infection in an alpine amphibian population. Viruses 11, 1–15.
Mitchell J., Vitikainen E. I. K., Wells D. A., Cant M. A. and Nichols H. J. 2017 Heterozygosity but not inbreeding coefficient predicts parasite burdens in the banded mongoose. J. Zool. 302, 32–39.
Monello R. J., Galloway N. L., Powers J. G., Madsen-Bouterse S. A., Edwards W. H. et al. 2017 Pathogen-mediated selection in free-ranging elk populations infected by chronic wasting disease. Proc. Natl. Acad. Sci.USA 114, 12208–12212.
Mooers A., Faith D. P. and Maddison W. P. 2008 Converting endangered species categories to probabilities of extinction for phylogenetic conservation prioritization. PLoS One 3, 1–5.
Moore J. 2002 Parasites and the behavior of animals, pp. 316. Oxford University Press. Ebook.
Morley N. J. and Lewis J. W. 2006 Anthropogenic pressure on a molluscan-trematode community over a long-term period in the Basingstoke Canal, UK, and its implications for ecosystem health. Ecohealth 3, 269–280.
Murray M. H., Becker D. J., Hall R. J. and Hernandez S. M. 2016 Wildlife health and supplemental feeding: a review and management recommendations. Biol. Conserv. 204, 163–174.
Nachev M. and Sures B. 2016 Environmental parasitology: parasites as accumulation bioindicators in the marine environment. J. Sea Res. 113, 45–50.
Newhouse D. J. and Balakrishnan C. N. 2015 High major histocompatibility complex class I polymorphism despite bottlenecks in wild and domesticated populations of the zebra finch (Taeniopygia guttata). BMC Evol. Biol. 15, 1–11.
Newman S. H., Hill N. J., Spragens K. A., Janies D., Voronkin I. O., Prosser D. J. et al. 2012 Eco-virological approach for assessing the role of wild birds in the spread of avian influenza H5N1 along the central Asian flyway. PLoS One 7, 1–12.
Nielsen R., Bustamante C., Clark A. G., Glanowski S., Sackton T. B., Hubisz M. J. et al. 2005 A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3, 0976–0985.
Niskanen A. K., Kennedy L. J., Ruokonen M., Kojola I., Lohi H., Isomursu M. et al. 2014 Balancing selection and heterozygote advantage in major histocompatibility complex loci of the bottlenecked Finnish wolf population. Mol. Ecol. 23, 875–889.
Nunes L. A., Turvey S. T. and Rosindell J. 2015 The price of conserving avian phylogenetic diversity: A global prioritization approach. Philos. Trans. R. Soc. B Biol. Sci. 370, 1–11.
O’Hanlon S. J., Rieux A., Farrer R. A., Rosa G. M., Waldman B. and Bataille A. 2018 Recent Asian origin of chytrid fungi causing global amphibian declines. Science 360, 621–627.
Ortego J., Aparicio J. M., Calabuig G. and Cordero P. J. 2007a Risk of ectoparasitism and genetic diversity in a wild lesser kestrel population. Mol. Ecol. 16, 3712–3720.
Ortego J., Cordero P. J., Aparicio J. M. and Calabuig G. 2007b No relationship between individual genetic diversity and prevalence of avian malaria in a migratory kestrel. Mol. Ecol. 16, 4858–4866.
Ostfeld R. S. 2013 A Candide response to Panglossian accusations by Randolph and Dobson: biodiversity buffers disease. Parasitology 140, 1196–1198.
Ostfeld R. S. and Logiudice K. 2003 Community disassembly, biodiversity loss, and the erosion of an ecosystem service. Ecology 84, 1421–1427.
Ostfeld R. S. and Keesing F. 2012 Effects of host diversity on infectious disease. Annu. Rev. Ecol. Evol. Syst. 43, 157–184.
Ostfeld R. S. and Keesing F. 2017 Is biodiversity bad for your health? Ecosphere 8.
Pafčo B., Čížková D., Kreisinger J., Hasegawa H., Vallo P., Shutt K. et al. 2018 Metabarcoding analysis of strongylid nematode diversity in two sympatric primate species. Sci. Rep. 8, 1–11.
Pagán I. and García-Arenal F. 2018 Tolerance to plant pathogens: Theory and experimental evidence. Int. J. Mol. Sci. 19, 810.
Panzera Y., Sarute N., Iraola G., Hernández M. and Pérez R. 2015 Molecular phylogeography of canine distemper virus: Geographic origin and global spreading. Mol. Phylogenet. Evol. 92, 147–154.
Paquette S. R., Talbot B., Garant D., Mainguy J. and Pelletier F. 2014 Modelling the dispersal of the two main hosts of the raccoon rabies variant in heterogeneous environments with landscape genetics. Evol. Appl. 7, 734–749.
Parker B. J., Garcia J. R. and Gerardo N. M. 2014 Genetic variation in resistance and fecundity tolerance in a natural host-pathogen interaction. Evolution 68, 2421–2429.
Pérez-del Olmo A., Raga J. A., Kostadinova A. and Fernández M. 2007 Parasite communities in Boops boops (L.) (Sparidae) after the Prestige oil-spill: Detectable alterations. Mar. Pollut. Bull. 54, 266–276.
Petren K. 2013 The evolution of landscape genetics. Evolution 67, 3383–3385.
Pinheiro L. B., Coleman V. A., Hindson C. M., Herrmann J., Hindson B. J., Bhat S. et al. 2012 Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 84, 1003–1011.
Poorten T. J. and Rosenblum E. B. 2016 Comparative study of host response to chytridiomycosis in a susceptible and a resistant toad species. Mol. Ecol. 25, 5663–5679.
Poulin R. 2010 Chapter 5 - Parasite manipulation of host behavior: an update and frequently asked questions. Adv. Study Behav. 41, 151–186.
Poulin R. and Morand S. 2000 The diversity of parasites. Q. Rev. Biol. 75, 277–293.
Pounds J. A., Bustamante M. R., Coloma L. A., Consuegra J. A., Fogden M. P. L. and Foster P. N. 2006 Widespread amphibian extinctions from epidemic disease driven by global warming. Nature 439, 161–167.
Prenter J., MacNeil C., Dick J. T. A. and Dunn A. M. 2004 Roles of parasites in animal invasions. Trends Ecol. Evol. 19, 385–390.
Preston D. L., Orlofske S. A., Lambden J. P. and Johnson P. T. J. 2013 Biomass and productivity of trematode parasites in pond ecosystems. J. Anim. Ecol. 82, 509–517.
Preston D. L., Jacobs A. Z., Orlofske S. A. and Johnson P. T. J. 2014 Complex life cycles in a pond food web: Effects of life stage structure and parasites on network properties, trophic positions and the fit of a probabilistic niche model. Oecologia 174, 953–965.
Preston D. L., Mischler J. A., Townsend A. R. and Johnson P. T. J. 2016 Disease ecology meets ecosystem science. Ecosystems 19, 737–748.
Price P. W., Westoby M., Rice B., Atsatt P. R., Fritz R. S., Thompson J. N. et al. 1986 Parasite mediation in ecological interactions. Annu. Rev. Ecol. Syst. 17, 487–505.
Prüter H., Franz M., Auls S., Czirják G., Greben O., Greenwood A. D. et al. 2018 Chronic lead intoxication decreases intestinal helminth species richness and infection intensity in mallards (Anas platyrhynchos). Sci. Total Environ. 644, 151–160.
Råberg L., Sim D. and Read A. F. 2007 Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science 318, 812–814.
Råberg L., Graham A. L. and Read A. F. 2009 Decomposing health: tolerance and resistance to parasites in animals. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 364, 37–49.
Ramírez J. D., Herrera G., Hernández C., Cruz-Saavedra L., Muñoz M., Flórez C et al. 2018 Evaluation of the analytical and diagnostic performance of a digital droplet polymerase chain reaction (ddPCR) assay to detect Trypanosoma cruzi DNA in blood samples. PLoS Negl. Trop. Dis. 12, e0007063.
Randolph S. E. and Dobson A. D. M. 2012 Pangloss revisited: a critique of the dilution effect and the biodiversity-buffers-disease paradigm. Parasitology 139, 847–863.
Rebollar E. A., Antwis R. E., Becker M. H., Belden L. K., Bletz M. C., Brucker R. M. et al. 2016 Using “omics” and integrated multi-omics approaches to guide probiotic selection to mitigate chytridiomycosis and other emerging infectious diseases. Front. Microbiol. 7, 68.
Reuter S., Connor T. R., Barquist L., Walker D., Feltwell T., Harris S. R. et al. 2014 Parallel independent evolution of pathogenicity within the genus Yersinia. Proc. Natl. Acad. Sci. USA 111, 6768–6773.
Richard F. A., Sehgal R. N. M., Jones H. I., Smith T. B. and Matbouli E. L. 2002 A comparative analysis of PCR-based detection methods for avian malaria. J. Parasitol. 88, 819–822.
Rijks J. M., Hoffman J. I., Kuiken T., Osterhaus A. D. M. E. and Amos W. 2008 Heterozygosity and lungworm burden in harbour seals (Phoca vitulina). Heredity 100, 587–593.
Riper C. van, Riper S. G. Van, Goff M. L. and Laird M. 1986 The epizootiology and ecological significance of malaria in Hawaiian land birds. Ecol. Monogr. 56, 327–344.
Rivero A. and Gandon S. 2018 Evolutionary ecology of avian malaria: past to present. Trends Parasitol. 34, 712–726.
Robar N., Burness G. and Murray D. L. 2010 Tropics, trophics and taxonomy: The determinants of parasite-associated host mortality. Oikos 119, 1273–1280.
Robin V. V., Gupta P., Thatte P. and Ramakrishnan U. 2015 Islands within islands: two montane palaeo-endemic birds impacted by recent anthropogenic fragmentation. Mol. Ecol. 24, 3572–3584.
Rodríguez-Ruano S. M., Škochová V., Rego R. O. M., Schmidt J. O., Roachell W., Hypša V. and Nováková E. 2018 Microbiomes of North American triatominae: The grounds for Chagas disease epidemiology. Front. Microbiol. 9, 1–11.
Rogalski M. A., Gowler C. D., Shaw C. L., Hufbauer R. A. and Duffy M. A. 2017 Human drivers of ecological and evolutionary dynamics in emerging and disappearing infectious disease systems. Philos. Trans. R. Soc. B Biol. Sci. 372, 20160043.
Rosenblum E. B., James T. Y., Zamudio K. R., Poorten T. J., Ilut D., Rodriguez D. et al. 2013 Complex history of the amphibian-killing chytrid fungus revealed with genome resequencing data. Proc. Natl. Acad. Sci. USA 110, 9385–9390.
Ruess R. W., McFarland J. M., Trummer L. M. and Rohrs-Richey J. K. 2009 Disease-mediated declines in N-fixation inputs by Alnus tenuifolia to early-successional floodplains in interior and south-central Alaska. Ecosystems 12, 489–502.
Ruiz-López M. J., Monello R. J., Gompper M. E. and Eggert L. S. 2012 The effect and relative importance of neutral genetic diversity for predicting parasitism varies across parasite taxa. PLoS One 7, e45404.
Rushton S. P., Lurz P. W. W., Gurnell J., Fuller R., Society B. E. and Ecology A. 2000 Modelling the spatial dynamics of parapoxvirus disease in red and grey squirrels: a possible cause of the decline in the red squirrel in the UK? J. Appl. Ecol. 37, 997–1012.
Salkeld D. J., Padgett K. A. and Jones J. H. 2013 A meta-analysis suggesting that the relationship between biodiversity and risk of zoonotic pathogen transmission is idiosyncratic. Ecol. Lett. 16, 679–686.
Sánchez C. A., Becker D. J., Teitelbaum C. S., Barriga P., Brown L. M., Majewska A. A. et al. 2018 On the relationship between body condition and parasite infection in wildlife: a review and meta-analysis. Ecol. Lett. 21, 1869–1884.
Sato C. F. and Lindenmayer D. B. 2018 Meeting the global challenge. Conserv. Lett. 11, 1–7.
Sato T., Watanabe K., Kanaiwa M., Niizuma Y., Harada Y. and Lafferty K. D. 2011 Nematomorph parasites drive energy flow through a riparian ecosystem. Ecology 92, 201–207.
Savage A. E. and Zamudio K. R. 2016 Adaptive tolerance to a pathogenic fungus drives major histocompatibility complex evolution in natural amphibian populations. Proc. R. Soc. B Biol. Sci. 283, 20153115.
Schneider D. S. and Ayres J. S. 2008 Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat. Rev. Immunol. 8, 889–895.
Schneider J., Valentini A., Dejean T., Montarsi F., Taberlet P., Glaizot O. et al. 2016 Detection of invasive mosquito vectors using environmental DNA (eDNA) from water samples. PLoS One 11, e0162493.
Schnell I. B., Sollmann R., Calvignac-Spencer S., Siddall M. E., Yu D. W., Wilting A. et al. 2015 iDNA from terrestrial haematophagous leeches as a wildlife surveying and monitoring tool - prospects, pitfalls and avenues to be developed. Front. Zool. 12, 1–14.
Schradin C., Lindholm A. K., Johannesen J., Schoepf I., Yuen C. H., König B. et al. 2012 Social flexibility and social evolution in mammals: a case study of the African striped mouse (Rhabdomys pumilio). Mol. Ecol. 21, 541–553.
Schwabl P., Llewellyn M., Landguth E. L. Andersson B., Kitron U., Costales J. A. et al. 2017 Prediction and prevention of parasitic diseases using a landscape genomics framework. Trends Parasitol. 33, 264–275.
Schwensow N., Mazzoni C. J., Marmesat E., Fickel J., Peacock D., Kovaliski J. et al. 2017 High adaptive variability and virus-driven selection on major histocompatibility complex (MHC) genes in invasive wild rabbits in Australia. Biol. Invasions 19, 1255–1271.
Scolari F., Casiraghi M. and Bonizzoni M. 2019 Aedes spp. and their microbiota: a review. Front. Microbiol. 10, 2036.
Selbach C., Jorge F., Dowle E., Bennett J., Chai X., Doherty J.-F. et al. 2019 Parasitological research in the molecular age. Parasitology 146, 1361–1370.
Sengupta M. E., Hellström M., Kariuki H. C., Olsen A., Thomsen P. F., Mejer H. et al. 2019 Environmental DNA for improved detection and environmental surveillance of schistosomiasis. Proc. Natl. Acad. Sci. USA 116, 8931–8940.
Sepil I., Lachish S., Hinks A. E. and Sheldon B. C. 2013 Mhc supertypes confer both qualitative and quantitative resistance to avian malaria infections in a wild bird population.
Seppälä O. and Jokela J. 2008 Host manipulation as a parasite transmission strategy when manipulation is exploited by non-host predators. Biol. Lett. 4, 663–666.
Shultz A. J. and Sackton T. B. 2019 Immune genes are hotspots of shared positive selection across birds and mammals. Elife 8, e41815.
Simms E. L. and Triplett J. 1994 Costs and benefits of plant responses to disease: resistance and tolerance. Evolution 48, 1973–1985.
Sin Y. W., Annavi G., Dugdale H. L., Newman C., Burke T. and MacDonald D. W. 2014 Pathogen burden, co-infection and major histocompatibility complex variability in the European badger (Meles meles). Mol. Ecol. 23, 5072–5088.
Skerratt L. F., Berger L., Speare R., Cashins S., McDonald K. R., Phillott A. D. et al. 2007 Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. Ecohealth 4, 125–134.
Smith K. F., Acevedo-Whitehouse K. and Pedersen A. B. 2009 The role of infectious diseases in biological conservation. Anim. Conserv. 12, 1–12.
Soares M. P., Teixeira L. and Moita L. F. 2017 Disease tolerance and immunity in host protection against infection. Nat. Rev. Immunol. 17, 83–96.
Sorensen A., van Beest F. M. and Brook R. K. 2013 Impacts of wildlife baiting and supplemental feeding on infectious disease transmission risk: A synthesis of knowledge. Prev. Vet. Med. 11, 1–8.
Soulé M. E. 1985 What is conservation biology? Bioscience 35, 727–734.
Stadler T. and Bonhoeffer S. 2013 Uncovering epidemiological dynamics in heterogeneous host populations using phylogenetic methods. Philos. Trans. R. Soc. B Biol. Sci. 368, 20120198.
Stevenson B., Goltz J. and Massé A. 2016 Preparing for and responding to recent incursions of racoon rabies variant into Canada. Canada Commun. Dis. Rep. 42, 125–129.
Stowe K. A., Marquis R. J., Hochwender C. G. and Simms E. L. 2000 The evolutionary ecology of tolerance to consumer damage. Annu. Rev. Ecol. Syst. 31, 565–595.
Stucki D., Ballif M., Bodmer T., Coscolla M., Maurer A. M., Droz S. et al. 2015 Tracking a tuberculosis outbreak over 21 years: Strain-specific single-nucleotide polymorphism typing combined with targeted whole-genome sequencing. J. Infect. Dis. 211,1306–1316.
Sures B. 2001 The use of fish parasites as bioindicators of heavy metals in aquatic ecosystems: A review. Aquat. Ecol. 35, 245–255.
Sures B. 2008 Environmental parasitology. Interactions between parasites and pollutants in the aquatic environment. Parasite 15, 434–438.
Sures B., Nachev M., Pahl M., Grabner D. and Selbach C. 2017a Parasites as drivers of key processes in aquatic ecosystems: Facts and future directions. Exp. Parasitol. 180, 141–147.
Sures B., Nachev M., Selbach C. and Marcogliese D. J. 2017b Parasite responses to pollution: what we know and where we go in ‘Environmental Parasitology.’ Parasit. Vectors 10, 1–19.
Sutton J. T., Castro I., Robertson B. C., Tompkins D. M., Stanton J. A. L. and Jamieson I. G. 2016 MHC genetic diversity and avian malaria prevalence in Mokoia Island saddlebacks. N. Z. J. Ecol. 40, 351–360.
Swetnam D., Widen S. G., Wood T. G., Reyna M., Wilkerson L., Debboun M. et al. 2018 Terrestrial bird migration and west nile virus circulation, United States. Emerg. Infect. Dis. 24, 2184–2194.
Taberlet P., Coissac E., Hajibabaei M. and Rieseberg L. H. 2012 Environmental DNA. Mol. Ecol. 21, 1789–1793.
Tanaka R., Hino A., Tsai I. J., Palomares-Rius J. E., Yoshida A., Ogura Y. et al. 2014 Assessment of helminth biodiversity in wild rats using 18S rDNA based metagenomics. PLoS One 9, e110769.
Telleria E. L., Martins-Da-Silva A., Tempone A. J. and Traub-Cseko Y. M. 2018 Leishmania, microbiota and sand fly immunity. Parasitology 145, 1336–1353.
Tellez M. and Merchant M. 2015 Biomonitoring heavy metal pollution using an aquatic apex predator, the American Alligator, and its Parasites. PLoS One 10, 1–18.
Teotônio I. M. S. N., Dias N., Hagström-Bex L., Nitz N., Francisco A. F. and Hecht M. 2019 Intestinal microbiota – A modulator of the Trypanosoma cruzi-vector-host triad. Microb. Pathog. 103711.
Thézé J., Li T., du Plessis L., Bouquet J., Kraemer M. U. G., Somasekar S. et al. 2018 Genomic epidemiology reconstructs the introduction and spread of Zika Virus in central America and Mexico. Cell Host Microbe 23, 855–864.e7.
Thogmartin W. E., Sanders-Reed C. A., Szymanski J. A., McKann P. C., Pruitt L. and King R. A. 2013 White-nose syndrome is likely to extirpate the endangered Indiana bat over large parts of its range. Biol. Conserv. 160, 162–172.
Thomas F., Schmidt-Rhaesa A., Martin G., Manu C., Durand P. and Renaud F. 2002 Do hairworms (Nematomorpha) manipulate the water seeking behaviour of their terrestrial hosts? J. Evol. Biol. 15, 356–361.
Thomsen P. F. and Willerslev E. 2015 Environmental DNA - An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18.
Tompkins D. M. and Gleeson D. M. 2006 Relationship between avian malaria distribution and an exotic invasive mosquito in new zealand. J. R. Soc. New Zeal. 36, 51–62.
Tompkins D. M., Sainsbury A. W., Nettleton P., Buxton D. and Gurnell J. 2002 Parapoxvirus causes a deleterious disease in red squirrels associated with UK population declines. Proc. R. Soc. B Biol. Sci. 269, 529–533.
Tompkins D. M., White A. R. and Boots M. 2003 Ecological replacement of native red squirrels by invasive greys driven by disease. Ecol. Lett. 6, 189–196.
Tompkins D. M., Dunn A. M., Smith M. J. and Telfer S. 2011 Wildlife diseases: From individuals to ecosystems. J. Anim. Ecol. 80, 19–38.
Torchin M. E., Lafferty K. D., Dobson A. P., McKenzie V. J. and Kuris A. M. 2003 Introduced species and their missing parasites. Nature 421, 628–630.
Torchin M. E., Lafferty K. D. and Kuris A. M. 2001 Release from parasites as natural enemies: Increased performance of a globally introduced marine crab. Biol. Invasions 3, 333–345.
Townsend A. K., Clark A. B., McGowan K. J., Buckles E. L., Miller A. D. and Lovette I. J. 2009 Disease-mediated inbreeding depression in a large, open population of cooperative crows. Proc. R. Soc. B 276, 2057–2064.
Townsend A. K., Taff C. C., Wheeler S. S., Weis A. M., Hinton M. G., Jones M. L. et al. 2018 Low heterozygosity is associated with vector-borne disease in crows. Ecosphere 9, e02407.
Trewby H., Nadin-Davis S. A., Real L. A. and Biek R. 2017 Processes underlying rabies virus incursions across US–Canada border as revealed by whole-genome phylogeography. Emerg. Infect. Dis. 23, 1454–1461.
Tschirren B. Andersson M., Scherman K., Westerdahl H., Mittl P. R. and Raberg L. 2013 Polymorphisms at the innate immune receptor TLR2 are associated with Borrelia infection in a wild rodent population. Proc. Biol. Sci. 280, 20130364.
Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R. and Gordon J. I. 2007 The Human Microbiome Project. Nature 449, 804–810.
Turner A. K., Begon M., Jackson J. A. and Paterson S. 2012 Evidence for selection at cytokine loci in a natural population of field voles (Microtus agrestis). Mol. Ecol. 21, 1632–1646.
Valkiŭnas G. 2005 Avian malaria parasites and other haemosporidia. Syst. Biol. 54, 860–863.
VanderWaal K. L., Atwill E. R., Isbell L. A. and McCowan B. 2014a Linking social and pathogen transmission networks using microbial genetics in giraffe (Giraffa camelopardalis). J. Anim. Ecol. 83, 406–414.
VanderWaal K. L., Atwill E. R., Isbell L. A. and McCowan B. 2014b Quantifying microbe transmission networks for wild and domestic ungulates in Kenya. Biol. Conserv. 169, 136–146.
Vannatta J. T. and Minchella D. J. 2018 Parasites and their impact on ecosystem nutrient cycling. Trends Parasitol. 34, 452–455.
Vidal-Martínez V. M., Pech D., Sures B., Purucker S. T. and Poulin R. 2010 Can parasites really reveal environmental impact? Trends Parasitol. 26, 44–51.
Videvall E., Cornwallis C. K., Palinauskas V., Valkiunas G. and Hellgren O. 2015 The avian transcriptome response to malaria infection. Mol. Biol. Evol. 32, 1255–1267.
Vilaça S. T., Grant S. A., Beaty L., Brunetti C. R., Congram M., Murray D. L. et al. 2019 Detection of spatiotemporal variation in ranavirus distribution using eDNA. Environ. DNA 1–11.
Volz E. M., Koelle K. and Bedford T. 2013 Viral Phylodynamics. PLoS Comput. Biol. 9, e1002947.
Volz E. M., Romero-Severson E. and Leitner T. 2017 Phylodynamic inference across epidemic scales. Mol. Biol. Evol. 34, 1276–1288.
Von Schantz T., Wittzell H., Goransson G., Grahn M. and Persson K. 1996 MHC genotype and male ornamentation: Genetic evidence for the Hamilton-Zuk model. Proc. R. Soc. B Biol. Sci. 263, 265–271.
Vrancken B., Lemey P., Rambaut A., Bedford T., Longdon B., Günthard H. F. et al. 2015 Simultaneously estimating evolutionary history and repeated traits phylogenetic signal: Applications to viral and host phenotypic evolution. Methods Ecol. Evol. 6, 67–82.
Vrijenhoek R. C. and Leberg P. L. 1991 Let’s not throw the baby out with the bathwater: a comment on management for mhc diversity in captive populations. Conserv. Biol. 5, 252–254.
Wang S., Liu C., Wilson A. B., Zhao N., Li X., Zhu W. et al. 2017 Pathogen richness and abundance predict patterns of adaptive major histocompatibility complex variation in insular amphibians. Mol. Ecol. 26, 4671–4685.
Warner R. E. 1968 The role of introduced diseases in the extinction of the endemic Hawaiian avifauna. Condor. 70, 101–120.
Weeks A. R., Turelli M., Harcombe W. R., Reynolds K. T. and Hoffmann A. A. 2007 From parasite to mutualist: Rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol. 5, 0997–1005.
Wegner K. M., Reusch T. B. H. and Kalbe M. 2003 Multiple parasites are driving major histocompatibility complex polymorphism in the wild. J. Evol. Biol. 16, 224–232.
Weinstein S., Titcomb G., Agwanda B., Riginos C. and Young H. 2017 Parasite responses to large mammal loss in an African savanna. Ecology 98, 1839–1848.
Weldon C., Du Preez L. H., Hyatt A. D., Muller R. and Speare R. 2004 Origin of the amphibian chytrid fungus. Emerg. Infect. Dis. 10, 2100.
Westermann A. J., Gorski S. A. and Vogel J. 2012 Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 10, 618–630.
Whiles M. R., Hall R. O., Dodds W. K., Verburg P., Huryn A. D. and Pringle C. M. 2012 Disease-Driven Amphibian Declines Alter Ecosystem Processes in a Tropical Stream. Ecosystems 16, 146–157.
White L. A., Forester J. D. and Craft M. E. 2017 Using contact networks to explore mechanisms of parasite transmission in wildlife. Biol. Rev. 92, 389–409.
Whitehorn P. R., Tinsley M. C., Brown M. J. F., Darvill B. and Goulson D. 2011 Genetic diversity, parasite prevalence and immunity in wild bumblebees. Proc. R. Soc. B Biol. Sci. 278, 1195–1202.
Wilder S. M., Raubenheimer D. and Simpson S. J. 2016 Moving beyond body condition indices as an estimate of fitness in ecological and evolutionary studies. Funct. Ecol. 30, 108–115.
Wilmers C. C., Post E., Peterson R. O. and Vucetich J. A. 2006 Predator disease out-break modulates top-down, bottom-up and climatic effects on herbivore population dynamics. Ecol. Lett. 9, 383–389.
Wilson M., Glaser K. C., Adams-Fish D., Boley M., Mayda M. and Molestina R. E. 2015 Development of droplet digital PCR for the detection of Babesia microti and Babesia duncani. Exp. Parasitol. 149, 24–31.
Winter M., Devictor V. and Schweiger O. 2013 Phylogenetic diversity and nature conservation: Where are we? Trends Ecol. Evol. 28, 199–204.
Winternitz J. C., Minchey S. G., Garamszegi L. Z., Huang S., Stephens P. R. and Altizer S. 2013 Sexual selection explains more functional variation in the mammalian major histocompatibility complex than parasitism. Proc. R. Soc. B Biol. Sci. 280, 20131605.
Wood C. L. and Johnson P. T. J. 2015 A world without parasites: exploring the hidden ecology of infection. Front. Ecol. Environ. 13, 425–434.
Woodworth B. L., Atkinson C. T., LaPointe D. A., Hart P. J., Spiegel C. S., Tweed E. J. et al. 2005 Host population persistence in the face of introduced vector-borne diseases: Hawaii amakihi and avian malaria. Proc. Natl. Acad. Sci. USA 102, 1531–1536.
Worby C. J., Lipsitch M. and Hanage W. P. 2017 Shared Genomic Variants: Identification of Transmission Routes Using Pathogen Deep-Sequence Data. Am. J. Epidemiol. 186, 1209–1216.
Wylie T. N., Wylie K. M., Herter B. N., Storch G. A., Author C. and Louis S. 2015 Enhanced Virome Sequencing Using Targeted Sequence Capture Enhanced virome sequencing using sequence capture. Genome Res. 4, 1910–1920.
Yap T. A., Nguyen N. T., Serr M., Shepack A. and Vredenburg V. T. 2017 Batrachochytrium salamandrivorans and the Risk of a Second Amphibian Pandemic. Ecohealth 14, 851–864.
Zhang L., Wu Q., Hu Y., Wu H. and Wei F. 2015 Major histocompatibility complex alleles associated with parasite susceptibility in wild giant pandas. Heredity 114, 85–93.
Acknowledgements
We thank colleagues at SREL for help and support. Financial support was provided from the US Department of Energy Financial Assistance Award no. DE-EM0004391 to the University of Georgia Research Foundation. VVR was supported by intramural funds from Indian Institute of Science Education and Research (IISER) Tirupati. Figures in the manuscript were created in Mind the Graph and Biorender platforms.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: T. N. C. Vidya
†This is one of the articles of collections on ‘Conservation Genetics’.
Appendix
Appendix
Parasite
Any organism that is associated with another organism (referred as the host) for all or part of its life cycle and acquires resources from the host, thereby reducing host fitness.
Pathogen
Parasites that cause disease in the host organism upon infection are referred to as pathogens.
Macroparasites and microparasites
Parasites can be classified as microparasites (bacteria, viruses, protozoa and fungi) and macroparasites (helminths and arthropods) based on their size and reproductive strategies. Microparasites are smaller with shorter generation times, reproduce intracellularly and multiply within the host. They are transmitted either by direct contact between hosts or via vector (usually an insect vector) and can also be transmitted vertically—from parents to offspring. In contrast, macroparasites are relatively larger with longer generation times and do not undergo direct reproduction or multiplication within the host. They are transmitted by the release of infective stages in the environment that are either directly taken up by another host or by predation upon an infected host.
Predation, herbivory and parasitism
Predation can simply be defined as consumption of one organism (the prey) by another organism (the predator), often in its entirety. Herbivory refers to the consumption of plant parts by animals (herbivores). Although parasitism is similar to other consumer–resource interactions such as predation and herbivory, there are ecological differences between them. One such dichotomy is whether the enemy completely eliminates its victim– a criterion almost universally accepted as central to predation but not always true of parasites. Parasites may reduce host fitness but do not always kill their hosts. However, some parasites (e.g. trophically transmitted parasites) could kill their host to facilitate their transmission onto the next host. Second, unlike predators, parasites are much smaller than their hosts. Third, predators can kill many diverse prey individuals throughout their lifetime; however, in contrast, parasites are restricted to one or few hosts.
Rights and permissions
About this article
Cite this article
Gupta, P., Robin, V.V. & Dharmarajan, G. Towards a more healthy conservation paradigm: integrating disease and molecular ecology to aid biological conservation†. J Genet 99, 65 (2020). https://doi.org/10.1007/s12041-020-01225-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-020-01225-7