Abstract
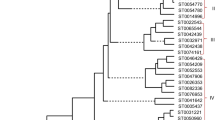
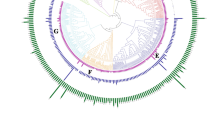
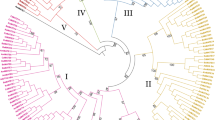
Multidrug and toxic compound extrusion (MATE) proteins are a group of secondary active transporters, which widely exist in all living organisms and play important role in the detoxication of endogenous secondary metabolites and exogenous agents. However, to date, no systematic and comprehensive study of this family is reported in maize. Here, a total of 49 MATE genes (ZmMATE) were identified and divided into seven groups by phylogenetic analysis. Conserved intro–exon structures and motif compositions were investigated in these genes. Results by gene locations indicated that these genes were unevenly distributed among all 10 chromosomes. Tandem and segmental duplications appeared to contribute to the expansion and evolution of this gene family. The K a/ K s ratios suggested that the ZmMATE has undergone large-scale purifying selection on the maize genome. Interspecies microsynteny analysis revealed that there were independent gene duplication events of 10 ZmMATE. In addition, most maize MATE genes exhibited different expression profiles in diverse tissues and developmental stages. Sixteen MATE genes were chosen for further quantitative real-time polymerase chain reaction analysis showed differential expression patterns in response to aluminum treatment. These results provide a useful clue for future studies on the identification of MATE genes and functional analysis of MATE proteins in maize.







Similar content being viewed by others
References
Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L. et al. 2009 MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208.
Cannon S. B., Mitra A., Baumgarten A., Young N. D. and May G. 2004 The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10.
Chen Y., Hao X. and Cao J. 2014 Small auxin upregulated RNA (SAUR) gene family in maize: Identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum . J. Int. Plant Biol. 56, 133–150.
Debeaujon I., Peeters A., Leon-Kloosterziel K. M. and Koornneef M. 2001 The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13, 853–871.
Delhaize E., Gruber B. D. and Ryan P. R. 2007 The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Lett. 581, 2255–2262.
Diener A. C., Gaxiola R. A. and Fink G. R. 2001 Arabidopsis ALF5, a multidrug efflux transporter gene family member, confers resistance to toxins. Plant Cell 13, 1625–1637.
Fan K., Wang M., Miao Y., Ni M., Bibi N., Yuan S. et al. 2014 Molecular evolution and expansion analysis of the NAC transcription factor in Zea mays. PLoS One 9, e111837.
Fujii M., Yokosho K., Yamaji N., Saisho D., Yamane M., Takahashi H. et al. 2012 Acquisition of aluminium tolerance by modification of a single gene in barley. Nat. Commun. 3, 713.
Furukawa J., Yamaji N., Wang H., Mitani N., Murata Y., Sato K. et al. 2007 An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 48, 1081–1091.
Gabelman W. H., Loughman B. C., Magnavaca R., Gardner C. O. and Clark R. B. 1987 Evaluation of inbred maize lines for aluminum tolerance in nutrient solution. In Genetic aspects of plant mineral nutrition (ed. W. H. Gabelman and B. C. Loughman), pp. 255–265. Springer, Netherlands.
Gallie D. R., Geisler-Lee J., Chen J. and Jolley B. 2009 Tissue-specific expression of the ethylene biosynthetic machinery regulates root growth in maize. Plant Mol. Biol. 69, 195–211.
Gaut B. S. 2002 Evolutionary dynamics of grass genomes. New Phytol. 154, 15–28.
Hvorup R. N., Winnen B., Chang A. B., Jiang Y., Zhou X. and Saier M. H. 2003 The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur. J. Biochem. 270, 799–813.
Ingram G. C., Boisnard-Lorig C., Dumas C. and Rogowsky P. M. 2000 Expression patterns of genes encoding HD-ZipIV homeo domain proteins define specific domains in maize embryos and meristems. Plant J. 22, 401–414.
Kochian L. V., Hoekenga O. A. and Piñeros M. A. 2004 How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorus efficiency. Annu. Rev. Plant Biol. 55, 459–493.
Koonin E. V. 2005 Orthologs, paralogs, and evolutionary genomics. In Annual review of genetics, pp. 309–338. Annual Reviews, Palo Alto.
Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D. et al. 2009 Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645.
Leng X., Liu D., Zhao M., Sun X., Li Y., Mu Q. et al. 2014 Genome-wide identification and analysis of FK506-binding protein family gene family in strawberry (Fragaria × ananassa). Gene 534, 390–399.
Li L. G., He Z. Y., Pandey G. K., Tsuchiya T. and Luan S. 2002 Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 277, 5360–5368.
Librado P. and Rozas J. 2009 DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452.
Liu J., Magalhaes J. V., Shaff J. and Kochian L. V. 2009 Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57, 389–399.
Livak K. J. and Schmittgen T. D. 2001 Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔCT method. Methods 25, 402–408.
Lynch M. and Conery J. S. 2003 The evolutionary demography of duplicate genes. J. Struct. Funct. Genomics 3, 35–44.
Magalhaes J. V., Liu J., Guimarães C. T., Lana U. G. P., Alves V. M. C., Wang Y. et al. 2007 A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 39, 1156–1161.
Maron L. G., Piñeros M. A., Guimarães C. T., Magalhaes J. V., Pleiman J. K., Mao C. et al. 2010 Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J. 61, 728–740.
Maron L. G., Guimaraes C. T., Kirst M., Albert P. S., Birchler J. A., Bradbury P. J. et al. 2013 Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc. Natl. Acad. Sci. USA 110, 5241–5246.
Morita Y., Kodama K., Shiota S., Mine T., Kataoka A., Mizushima T. et al. 1998 NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42, 1778–1782.
Omote H., Hiasa M., Matsumoto T., Otsuka M. and Moriyama Y. 2006 The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 27, 587–593.
Peng X., Zhao Y., Cao J., Zhang W., Jiang H., Li X. et al. 2012 CCCH-type zinc finger family in maize: genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS One 7, e40120.
Pickett F. B. and Meeks-Wagner D. R. 1995 Seeing double: appreciating genetic redundancy. Plant Cell 7, 1347–1356.
Piñeros M. A., Magalhaes J. V., Carvalho Alves V. M. and Kochian L. V. 2002 The physiology and biophysics of an aluminum tolerance mechanism based on root citrate exudation in maize. Plant Physiol. 129, 1194–1206.
Ryan P. R., Raman H., Gupta S., Horst W. J. and Delhaize E. 2009 A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots. Plant Physiol. 149, 340–351.
Salse J., Bolot S., Throude M., Jouffe V., Piegu B., Quraishi U. M. et al. 2008 Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20, 11–24.
Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., Pasternak S. et al. 2009 The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115.
Sekhon R. S., Lin H., Childs K. L., Hansey C. N., Buell C. R., Leon N. et al. 2011 Genome-wide atlas of transcription during maize development. Plant J. 66, 553–563.
Sivaguru M., Liu J. and Kochian L. V. 2013 Targeted expression of SbMATE in the root distal transition zone is responsible for sorghum aluminum resistance. Plant J. 76, 297–307.
Swigoňová Z., Lai J. S., Ma J. X., Ramakrishna W., Llaca V., Bennetzen J. L. et al. 2004 Close split of sorghum and maize genome progenitors. Genome Res. 14, 1916–1923.
Takanashi K., Yokosho K., Saeki K., Sugiyama A., Sato S., Tabata S. et al. 2013 LjMATE1: a citrate transporter responsible for iron supply to the nodule infection zone of lotus japonicas. Plant Cell Physiol. 54, 1749.
Takanashi K., Shitan N. and Yazaki K. 2014 The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotechnol. 31, 417–430.
Tamura K., Peterson D., Peterson N., Stecher G., Nei M. and Kumar S. 2011 MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Taylor J. S. and Raes J. 2004 Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 38, 615–643.
Tiwari M., Sharma D., Singh M., Tripathi R. D. and Trivedi P. K. 2014 Expression of OsMATE1 and OsMATE2 alters development, stress responses and pathogen susceptibility in Arabidopsis. Sci. Rep. 4, 3964.
Vision T. J., Brown D. G. and Tanksley S. D. 2000 The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117.
Vonuexkull H. R. and Mutert E. 1995 Global extent, development and economic impact of acid soils. Plant Soil 171, 1–15.
Wang Y., Tang H., DeBarry J. D., Tan X., Li J., Wang X. et al. 2012 MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49.
Wei F., Coe E., Nelson W., Bharti A. K., Engler F., Butler E. et al. 2007 Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 3, 1254–1263.
Yang Z., Gu S., Wang X., Li W., Tang Z. and Xu C. 2008 Molecular evolution of the CPP-like gene family in plants: Insights from comparative genomics of Arabidopsis and rice. J. Mol. Evol. 67, 266–277.
Yokosho K., Yamaji N. and Ma J. F. 2010 Isolation and characterisation of two MATE genes in rye. Funct. Plant Biol. 37, 296–303.
Acknowledgements
This study was supported by the key project of National Natural Science Foundation of China (31501321) the Anhui University Natural Science Research Projects (KJ2015A100) and Anhui Postdoctoral Science Foundation and Colleges. We would like to thank the members of the Key Laboratory of Crop Biology of Anhui province for their assistance in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Zhu H., Wu J., Jiang Y., Jin J., Zhou W., Wang Y., Han G., Zhao Y. and Cheng B. 2016 Genomewide analysis of MATE-type gene family in maize reveals microsynteny and their expression patterns under aluminum treatment. J. Genet. 95, xx–xx]
Huasheng Zhu and Jiandong Wu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
ZHU, H., WU, J., JIANG, Y. et al. Genomewide analysis of MATE-type gene family in maize reveals microsynteny and their expression patterns under aluminum treatment. J Genet 95, 691–704 (2016). https://doi.org/10.1007/s12041-016-0686-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-016-0686-2