Abstract
This article describes the basic principles of steady-state and time-resolved fluorescence. The formal equivalence of the two methodologies is described first, followed by the extra advantages of time-resolved methods in revealing population heterogeneity in complex systems encountered in biology. Several examples from the author’s work are described in support of the above contention. Finally, several misinterpretations and pitfalls in the interpretation of fluorescence data and their remedy are described.

Reproduced with permission from Swaminathan et al. (1996).

Figure reproduced with permission from Lakshmikanth et al. (2001).

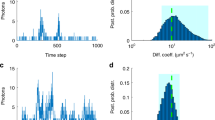
Reproduced with permission from Bhatia et al. (2018).

Reproduced with permission from Bhatia et al. (2018).


Reproduced with permission from Biswas et al. (2017).
Similar content being viewed by others
References
Alcala JR, Gratton E and Prendergast FG 1987 Fluorescence lifetime distribution in proteins. Biophys. J. 51 597–604
Bhatia S, Krishnamoorthy G and Udgaonkar JB 2018 Site-specific time-resolved FRET reveals local variations in the unfolding mechanism in an apparently two-state protein unfolding transition. Phys. Chem. Chem. Phys. 20 3216–3232
Biswas A, Mariam J, Kombrabail M, Narayan S, Krishnamoorthy G and Anand R 2017 Site-Specific Fluorescence Dynamics to Probe Polar Arrest by Fob1 in Replication Fork Barrier Sequences. ACS Omega 2 7389–7399
Brochon JC 1994 Maximum entropy method of data analysis in time-resolved spectroscopy. Methods Enzymol. 240 262–311
Bryngelson JD, Onuchic JN, Socci ND and Wolynes PG 1995 Funnels, pathways and the energy landscape of protein folding. Proteins 21 167–195
Chakraborty H, Haldar S, Lee-Gau Chong P, Kombrabail M, Krishnamoorthy G and Chattopadhyay A 2015 Depth-Dependent Organization and Dynamics of Archaeal and Eukaryotic Membranes: Development of Membrane Anisotropy Gradient with Natural Evolution. Langmuir 31 11591–11597
Das TK, Periasamy N and Krishnamoorthy G 1991 Mechanism of response of Potential-Sensitive dyes studied by Time-resolved fluorescence. Biophys. J. 64 1122–1132
Ebner TJ and Chen G 1995 Use of voltage-sensitive dyes and optical recordings in the central nervous system. Progress in Neurobiology 46 463–506
Ghosh S, Salot S, Sengupta S, Navalkar A, Ghosh D, Jacob R, Das S, Kumar R, Jha NN, Sahay S, Mehra S, Mohite GM, Ghosh SK, Kombrabail M, Krishnamoorthy G, Chaudhari P and Maji SK 2017 p53 amyloid formation leading to its loss of function: implications in cancer pathogenesis. Cell Death Differen. 24 1784–1798
Haldar S, Kombrabail M, Krishnamoorthy G and Chattopadhyay A 2010 Monitoring Membrane Protein Conformational Heterogeneity by Fluorescence Lifetime Distribution Analysis using the Maximum Entropy Method. J. Fluores. 20 407–413
Haldar S, Kombrabail M, Krishnamoorthy G and Chattopadhyay A 2012 Depth-Dependent Heterogeneity in Membranes by Fluorescence Lifetime Distribution Analysis. J. Phys. Chem. Lett. 3 2676–2681
Han J and Burgess K 2010 Fluorescent indicators for intracellular pH. Chem. Rev. 110 2709–2728
Hoong‐Sun Im H-S and Bernstein ER 1988 Geometry and torsional motion of biphenyl in the ground and first excited singlet state. J. Chem. Phys. 88 7337–7343
Jha SK, Dhar D, Krishnamoorthy G and Udgaonkar JB 2009a Continuous dissolution of structure during the unfolding of a small protein. Proc. Natl. Acad. Sci. USA 106 11113–11118
Jha A, Udgonakar JB and Krishnamoorthy G 2009b Characterization of the heterogeneity and specificity of inter-polypeptide interactions in amyloid protofibrils by measurement of site-specific fluorescence anisotropy decay kinetics. J. Mol. Biol. 393 735–752
Jha A, Ishii K, Udgaonkar JB, Tahara T and Krishnamoorthy G 2011 Exploration of the correlation between solvation dynamics and internal dynamics of a protein. Biochemistry 50 397–408
Jha A, Narayan S, Udgaonkar JB and Krishnamoorthy G 2012 Solvent-Induced Tuning of Internal Structure in a Protein Amyloid Protofibril. Biophys. J. 103 797–806
Jun JJ, Steinmetz NA and Harris TD 2017 Fully integrated silicon probes for high-density recording of neural activity. Nature 551 232–236
Krishnamoorthy G and Srivastava A 1997 Intracellular dynamics seen through time-resolved fluorescence microscopy. Curr. Sci. 72 835–845
Krishnamoorthy G and Ira 2001 Fluorescence Lifetime distribution in characterizing membrane microheterogeneity. J. Fluorescence 11 247–253
Krishnamoorthy G 2003 Fluorescence spectroscopy in molecular description of biological processes. Indian J. Biochem. Biophys. 40 147–159
Krishnamoorthy G 2012 Motional dynamics in proteins and nucleic acids control their function: revelation by time-domain fluorescence. Curr. Sci. 102 266–276
Lakowicz JR 2006 Principles of Fluorescence Spectroscopy (New York: Kluwer Academic/Plenum Publishers)
Lakshmikanth GS and Krishnamoorthy G 1999 Solvent-exposed tryptophans probe the dynamics at protein surfaces. Biophys. J. 77 1100–1106
Lakshmikanth GS, Sridevi K, Krishnamoorthy G and Udgaonkar JB 2001 Structure is lost incrementally during the unfolding of barstar. Nat. Struct. Biol. 8 799–804
Loubes J-M and Pelletier B 2008 Maximum entropy solution to ill-posed inverse problem with approximately known operator. J. Math. Anal. App. 344 260–273
Lyubovitsky JG, Gray HB and Winkler JR 2002 Mapping the cytochrome C folding landscape. J. Am. Chem. Soc. 124 5481–5485
Modrich P and Lahue R 1996 Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu. Rev. Biochem. 65 101–133
Mondal S, Kallianpur MV, Udgaonkar JB and Krishnamoorthy G 2016 Molecular crowding causes narrowing of population heterogeneity and restricts internal dynamics in a protein. Methods Appl. Fluorescence 4 014003
Mukherjee S, Kombrabail M, Krishnamoorthy G and Chattopadhyay A 2007 Dynamics and heterogeneity of bovine hippocampal membranes: Role of cholesterol and proteins. Biochim. Biophys. Acta 1768 2130–2144
Mukhopadhyay S, Nayak PK, Udgaonkar JB and Krishnamoorthy G 2006 Characterization of the formation of amyloid protofibrils from barstar by mapping residue-specific fluorescence dynamics. J. Mol. Biol. 358 935–942
Nag N, Krishnamoorthy G and Rao BJ 2005 A single mismatch in the dna induces enhanced aggregation of MutS: Hydrodynamic analysis of the protein–DNA complexes. FEBS J. 272 6228–6243
Nag N, Ramreddy T, Kombrabhail MH, Krishnamohan PM, D’souza J, Rao BJ, Duportail G, Mely Y and Krishnamoorthy G 2006 Dynamics of DNA and Protein–DNA Complexes Viewed Through Time-Domain Fluorescence; in Reviews in Fluorescence Vol. 3 (eds) C Geddes and J Lakowicz (New York: Springer Science+Business Media) pp 311–340
Nag N, Rao BJ and Krishnamoorthy G 2007 Altered dynamics of DNA bases adjacent to a mismatch: a cue for mismatch recognition by MutS. J. Mol. Biol. 374 39–53
Narayan S, Kombrabail MH, Das S, Singh H, Chary KVR, Rao BJ and Krishnamoorthy G 2015 Site-specific fluorescence dynamics in an RNA ‘thermometer’ reveals the role of ribosome binding in its temperature-sensitive switch function. Nucleic Acids Res. 43 493–503
Navon A, Ittah V, Landsman P, Scheraga HA and Haas E 2001 Distributions of intramolecular distances in the reduced and denatured states of bovine pancreatic ribonuclease A. Biochemistry 40 105–118
Paila Y, Kombrabhail M, Krishnamoorthy G and Chattopadhyay A 2011 Oligomerization of Serotonin 1A Receptor in Live Cells: A Time-Resolved Fluorescence Anisotropy Approach. J. Phys. Chem. B 115 11439–11447
Rami BR, Krishnamoorthy G and Udgaonkar JB 2003 Dynamics of the core tryptophan during the formation of a productive molten globule intermediate of barstar. Biochemistry 42 7986–8000
Ramreddy T, Sen S, Rao BJ and Krishnamoorthy G 2003 DNA Dynamics in RecA-DNA Filaments: ATP Hydrolysis-Related Flexibility in DNA. Biochemistry 42 12085–12094
Ramreddy T, Kombrabail M, Krishnamoorthy G and Rao BJ 2009 Site-specific dynamics in TAT triplex DNA as revealed by time-domain fluorescence of 2-Aminopurine. J. Phys. Chem. 113 6840–6846
Sahay S, Anoop A, Krishnamoorthy G and Maji SK 2014 Site-specific fluorescence dynamics of α-Synuclein fibrils using time-resolved fluorescence studies: effect of familial Parkinson’s disease-associated mutations. Biochemistry 53 807–809
Sahay S, Ghosh D, Dwivedi S, Anoop A, Kombrabail MH, Mohite GM, Krishnamoorthy G and Maji SK 2015 Familial Parkinson’s disease associated mutations alter the site-specific microenvironment and dynamics of α-Synuclein. J. Biol. Chem. 290 7804–7822
Sarkar SS, Saxena A, Dhawale N, Udgaonkar JB, and Krishnamoorthy G 2009 Protein folding, unfolding and aggregation process revealed by rapid sampling of time-domain fluorescence; in Reviews in Fluorescence (ed) CD Geddes (New York: Springer) pp 281–301
Saxena A, Udgaonkar JB and Krishnamoorthy G 2005 Protein dynamics and protein folding dynamics revealed by time-resolved fluorescence; in Fluorescence Spectroscopy in Biology, Advanced Methods and their Applications to Membranes, Proteins, DNA and Cells Springer Series on Fluorescence, Vol.3 (eds) Martin Hof, R Hutterer and V Fidler (Berlin: Springer) pp 163–179
Sekek O, Henry-Toulme N, Sureau F and Bolard J 1991 SNARF-1 as an intracellular pH-indicator in laser microspectrofluorimetry: A critical assessment. Anal. Biochem. 193 49–54
Sharma P, Varma R, Sarasij RC, Ira, Gousset K, Krishnamoorthy G, Rao M and Mayor S 2004 Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell 116 577–589
Sridevi K, Bhuyan A, Juneja J, Krishnamoorthy G and Udgaonkar JB 2000 The slow folding reaction of Barstar: the core tryptophan region attains tight packing before substantial secondary and tertiary structure formation and final compaction of the polypeptide chain. J. Mol. Biol 302 479–495
Srivastava A and Krishnamoorthy G 1997a Fluid-Phase Viscosity of cytoplasm estimated by time-resolved fluorescence microscopy. Archiv. Biochem. Biophys. 340 159–167
Srivastava A and Krishnamoorthy G 1997b Time-resolved Fluorescence Microscopy Corrects for the Probe Binding while Estimating Intracellular pH. Anal. Biochem. 249 140–146
Swaminathan R, Krishnamoorthy G and Periasamy N 1994a Similarity of fluorescence lifetime distributions for single tryptophan proteins in the random coil state. Biophys. J. 67 2013–2023
Swaminathan R, Periasamy N, Udgaonkar JB and Krishnamoorthy G 1994b Molten globule-like conformation of Barstar: a study by fluorescence dynamics. J. Phys. Chem. 98 9270–9278
Swaminathan R and Periasamy N 1996 Analysis of fluorescence decay by maximum entropy method: Influence of noise and analysis parameters on the width of the distribution of lifetimes. Proc. Indian Acad. Sci. (Chem. Sci. ) 108 39–49
Swaminathan R, Nath U, Udgaonkar JB, Periasamy N and Krishnamoorthy G 1996 Motional dynamics of a buried tryptophan reveals the presence of partially structured forms during denaturation of Barstar. Biochemistry 35 9150–9157
Tanaka F and Mataga M 1979 Theory of time-dependent photo selection in interacting fixed systems. Photochem. Photobiol. 29 1091–1097
Tsang E and Carr AM 2008 Replication fork arrest, recombination and the maintenance of ribosomal DNA stability. DNA Repair 7 1613−1623
Valeur B 2002 Molecular Fluorescence (Weinham: Wiley-VCH)
Weber G 1954 Dependence of polarization of the fluorescence on the concentration. Trans. Faraday Soc. 50 552–555
Yao S, Schafer-Hales KJ and Belfield KD 2007 A new water-soluble near-neutral ratiometric fluorescent pH indicator. Org. Lett. 9 5645–5648
Acknowledgements
Collaboration with Prof Jayant Udgaonkar, Prof Ruchi Anand, Prof R Swaminathan, Dr G Lakshmikanth, Dr K Sridevi, Dr Tapan Das, Dr Arvind Srivastava, Ms Sandhya Bhatia, Dr Anwesha Biswas and Dr Jessy Miriam John is gratefully acknowledged. Their work is described in some detail here. I thank Prof N Periasamy for providing the software used in the analysis of data shown here.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Krishnamoorthy, G. Fluorescence spectroscopy for revealing mechanisms in biology: Strengths and pitfalls. J Biosci 43, 555–567 (2018). https://doi.org/10.1007/s12038-018-9763-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12038-018-9763-4