Abstract
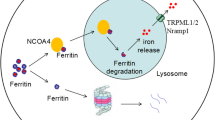
Mounting evidence indicates that the lysosome-autophagy pathway plays a critical role in iron release from ferritin, the main iron storage cellular protein, hence in the distribution of iron to the cells. The recent identification of nuclear receptor co-activator 4 as the receptor for ferritin delivery to selective autophagy sheds further light on the understanding of the mechanisms underlying this pathway. The emerging view is that iron release from ferritin through the lysosomes is a general mechanism in normal and tumour cells of different tissue origins, but it has not yet been investigated in brain cells. Defects in the lysosome-autophagy pathway are often involved in the pathogenesis of neurodegenerative disorders, and brain iron homeostasis disruption is a hallmark of many of these diseases. However, in most cases, it has not been established whether iron dysregulation is directly involved in the pathogenesis of the diseases or if it is a secondary effect derived from other pathogenic mechanisms. The recent evidence of the crucial involvement of autophagy in cellular iron handling offers new perspectives about the role of iron in neurodegeneration, suggesting that autophagy dysregulation could cause iron dyshomeostasis. In this review, we recapitulate our current knowledge on the routes through which iron is released from ferritin, focusing on the most recent advances. We summarise the current evidence concerning lysosome-autophagy pathway dysfunctions and those of iron metabolism and discuss their potential interconnections in several neurodegenerative disorders, such as Alzheimer’s, Parkinson’s and Huntington’s diseases; amyotrophic lateral sclerosis; and frontotemporal lobar dementia.




Similar content being viewed by others
Abbreviations
- AD:
-
Alzheimer’s disease
- ALAS2:
-
5′-Aminolevulinate synthase 2
- ALS:
-
Amyotrophic lateral sclerosis
- ALS2:
-
Amyotrophic lateral sclerosis 2, juvenile
- APs:
-
Amyloid plaques
- APP:
-
Amyloid precursor protein (amyloid beta A4 precursor protein)
- ATG5:
-
Autophagy related 5
- ATG7:
-
Autophagy related 7
- ATG9A:
-
Autophagy related 9A
- ATG11:
-
Autophagy related 11
- ATP13A2/PARK9:
-
ATPase type 13A2
- ATXN2:
-
Ataxin 2
- AV:
-
Autophagic vacuole
- BBB:
-
Blood-brain barrier
- C9orf72:
-
Chromosome 9 open reading frame 72
- C19orf12:
-
Chromosome 19 open reading frame 12
- CBD:
-
Corticobasal degeneration
- CHMP2B:
-
Charged multivesicular body protein 2B
- CMA:
-
Chaperone-mediated autophagy
- CoASY:
-
Coenzyme-A synthase
- CP:
-
Ceruloplasmin
- CTSC:
-
Cathepsin C
- DAO:
-
d-Aminoacid oxidase
- DCYTB:
-
Duodenal cytochrome b (CYBRD1, cytochrome b reductase 1)
- DMT1:
-
Divalent metal transporter 1 (SLC11A2, solute carrier family 11, proton-coupled divalent metal ion transporter, member 2)
- DJ-1/PARK7:
-
Parkinson protein 7
- FA2H:
-
Fatty acid 2 hydroxylase
- FBXL5:
-
F-box and leucine-rich repeat protein 5
- FPN1:
-
Ferroportin 1 (SLC40A1, solute carrier family 40, iron-regulated transporter, member 1)
- FT:
-
Ferritin
- FTH1:
-
Ferritin heavy polypeptide 1
- FTL:
-
Ferritin light polypeptide
- FTLD:
-
Frontotemporal lobar dementia
- FTMT:
-
Mitochondrial ferritin
- FRDA:
-
Friedreich’s ataxia
- FUS:
-
Fused in sarcoma (FUS RNA-binding protein)
- FXN:
-
Frataxin
- GABARAP:
-
GABA(A) receptor-associated protein
- GABARAPL1:
-
GABA(A) receptor-associated protein-like 1
- GABARAPL2:
-
GABA(A) receptor-associated protein-like 2
- GBA1:
-
Glucocerebrosidase A1 (GBA, glucosidase beta, acid)
- HAMP:
-
Hepcidin
- HCP1:
-
Heme carrier protein 1 (SLC46A1, solute carrier family 46, folate transporter, member 1)
- HD:
-
Huntington’s disease
- HEPH:
-
Hephaestin
- HF:
-
Hereditary ferritinopathy
- HFE:
-
Haemochromatosis
- HO1:
-
Heme oxygenase 1 (HMOX1)
- HTRA2/PARK13:
-
HtrA serine peptidase 2
- HTT:
-
Huntingtin
- IRE:
-
Iron-responsive element
- IRP1:
-
Iron regulatory protein 1 (ACO1, aconitase 1)
- IRP2:
-
Iron regulatory protein 2 (IREB2, iron-responsive element-binding protein 2)
- LAMP1:
-
Lysosomal-associated membrane protein 1
- LAMP2:
-
Lysosomal-associated membrane protein 2
- LC3:
-
Microtubule-associated protein 1 light chain 3 alpha (MAP1LC3A)
- LIP:
-
Labile iron pool
- LRRK2/PARK8:
-
Leucine-rich repeat kinase 2
- LSD:
-
Lysosomal storage disease
- MAPT:
-
Microtubule-associated protein tau
- MCOLN1:
-
Mucolipin 1
- MEF:
-
Mouse embryonic fibroblast
- MFN1:
-
Mitoferrin 1 (SLC25A37, solute carrier family 25 member 37)
- MFN2:
-
Mitoferrin 2 (SLC25A28, solute carrier family 25 member 28)
- MSA:
-
Multiple system atrophy
- mtor:
-
Mechanistic target of rapamycin
- NBIA:
-
Neurodegeneration with brain iron accumulation
- NBR1:
-
Neighbour of BRCA1 gene 1
- NCOA4:
-
Nuclear receptor co-activator 4
- NDP52:
-
Nuclear dot protein 52 (CALCOCO2, calcium binding and coiled-coil domain 2)
- NFT:
-
Neurofibrillary tangles
- NTBI:
-
Non-transferrin bound iron
- OPTN:
-
Optineurin
- PANK2:
-
Pantothenate kinase 2
- PARK2:
-
Parkin
- PCBP1:
-
Poly rC-binding protein 1
- PCBP2:
-
Poly rC-binding protein 2
- PD:
-
Parkinson’s disease
- PFN1:
-
Profilin 1
- PGRN:
-
Progranulin (GRN, granulin)
- PINK1/PARK6:
-
PTEN-induced putative kinase 1
- PLA2G6/PARK14:
-
Phospholipase A2 group VI
- PSEN1:
-
Presenilin 1
- PSENEN:
-
Presenilin enhancer γ-secretase subunit
- PSP:
-
Progressive supranuclear palsy
- RAB29/PARK16:
-
RAB29 member RAS oncogene family (PARK16, Parkinson disease 16)
- RAB38:
-
RAB38 member RAS oncogene family
- ROS:
-
Reactive oxygen species
- SG:
-
Stress granule
- SIGMAR1:
-
Sigma non-opioid intracellular receptor 1
- SILAC:
-
Stable isotope labelling by amino acids in cell culture
- SNCA:
-
α-Synuclein
- SNCA/PARK1:
-
α-Synuclein gene mutation
- SNCA/PARK4:
-
α-Synuclein gene multiplication
- SOD1:
-
Superoxide dismutase 1
- SOD2:
-
Superoxide dismutase 2, mitochondrial
- SQSTM1:
-
Sequestosome 1
- SREBF1:
-
Sterol regulatory element-binding transcription factor 1
- STEAP3:
-
STEAP family member 3, metalloreductase
- TDP-43:
-
TAR DNA-binding protein (TARDBP)
- TF:
-
Transferrin
- TFEB:
-
Transcription factor EB
- TFR1:
-
Transferrin receptor 1 (TFRC, transferrin receptor)
- TIM2:
-
T cell immunoglobulin and mucin domain-2 (Timd2)
- TMEM106B:
-
Transmembrane protein 106B
- TMPRSS6:
-
Transmembrane protease, serine 6
- UB:
-
Ubiquitin
- UBQLN2:
-
Ubiquilin 2
- UCH-L1/PARK5:
-
Ubiquitin carboxyl-terminal esterase L1
- UPS:
-
Ubiquitin proteasome system
- UTR:
-
Untranslated region
- VAPB:
-
VAMP-associated protein B and C
- VCP:
-
Valosin-containing protein
- VPS35/PARK17:
-
Vacuolar protein sorting 35 (VPS35 retromer complex component)
- WDR45:
-
WD repeat domain 45
References
Beard JL, Connor JR (2003) Iron status and neural functioning. Annu Rev Nutr 23:41–58
Muhoberac BB, Vidal R (2013) Abnormal iron homeostasis and neurodegeneration. Front Aging Neurosci 5:32
Kono S (2012) Aceruloplasminemia. Curr Drug Targets 13:1190–1199
Ward RJ, Zucca FA, Duyn JH, Crichton RR, Zecca L (2014) The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol 13:1045–1060
Linder MC (2013) Mobilization of stored iron in mammals: a review. Nutr 5:4022–4050
Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC (2014) Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509:105–109
Silva B, Faustino P (2015) An overview of molecular basis of iron metabolism regulation and the associated pathologies. Biochim Biophys Acta 1852:1347–1359
Lane DJ, Merlot AM, Huang ML, Bae DH, Jansson PJ, Sahni S, Kalinowski DS, Richardson DR (2015) Cellular iron uptake, trafficking and metabolism: key molecules and mechanisms and their roles in disease. Biochim Biophys Acta 1853:1130–1144
Theil EC (2011) Ferritin protein nanocages use ion channels, catalytic sites, and nucleation channels to manage iron/oxygen chemistry. Curr Opin Chem Biol 15:304–311
Merle U, Fein E, Gehrke SG, Stremmel W, Kulaksiz H (2007) The iron regulatory peptide hepcidin is expressed in the heart and regulated by hypoxia and inflammation. Endocrinology 148:2663–2668
Kulaksiz H, Fein E, Redecker P, Stremmel W, Adler G, Cetin Y (2008) Pancreatic beta-cells express hepcidin, an iron-uptake regulatory peptide. J Endocrinol 197:241–249
Bekri S, Gual P, Anty R, Luciani N, Dahman M, Ramesh B, Iannelli A, Staccini-Myx A et al (2006) Increased adipose tissue expression of hepcidin in severe obesity is independent from diabetes and NASH. Gastroenterology 131:788–796
Cho HH, Cahill CM, Vanderburg CR, Scherzer CR, Wang B, Huang X, Rogers JT (2010) Selective translational control of the Alzheimer amyloid precursor protein transcript by iron regulatory protein-1. J Biol Chem 285:31217–31232
Febbraro F, Giorgi M, Caldarola S, Loreni F, Romero-Ramos M (2012) α-Synuclein expression is modulated at the translational level by iron. Neuroreport 23:576–580
Rouault TA (2013) Iron metabolism in the CNS: implications for neurodegenerative diseases. Nat Rev Neurosci 14:551–564
Dickinson TK, Connor JR (1995) Cellular distribution of iron, transferrin, and ferritin in the hypotransferrinemic (Hp) mouse brain. J Comp Neurol 355:67–80
Todorich B, Zhang X, Slagle-Webb B, Seaman WE, Connor JR (2008) Tim-2 is the receptor for H-ferritin on oligodendrocytes. J Neurochem 107:1495–1505
Marques F, Falcao AM, Sousa JC, Coppola G, Geschwind D, Sousa N, Correia-Neves M, Palha JA (2009) Altered iron metabolism is part of the choroid plexus response to peripheral inflammation. Endocrinology 150:2822–2828
McCarthy RC, Kosman DJ (2015) Iron transport across the blood–brain barrier: development, neurovascular regulation and cerebral amyloid angiopathy. Cell Mol Life Sci 72:709–727
Zechel S, Huber-Wittmer K, von B, Halbach O (2006) Distribution of the iron-regulating protein hepcidin in the murine central nervous system. J Neurosci Res 84:790–800
Urrutia P, Aguirre P, Esparza A, Tapia V, Mena NP, Arredondo M, González-Billault C, Núñez MT (2013) Inflammation alters the expression of DMT1, FPN1 and hepcidin, and it causes iron accumulation in central nervous system cells. J Neurochem 126:541–549
Qian ZM, He X, Liang T, Wu KC, Yan YC, Lu LN, Yang G, Luo QQ et al (2014) Lipopolysaccharides upregulate hepcidin in neuron via microglia and the IL-6/STAT3 signaling pathway. Mol Neurobiol 50:811–820
Cocco E, Porrini V, Derosas M, Nardi V, Biasiotto G, Maccarinelli F, Zanella I (2013) Protective effect of mitochondrial ferritin on cytosolic iron dysregulation induced by doxorubicin in HeLa cells. Mol Biol Rep 40:6757–6764
Simpson IA, Ponnuru P, Klinger ME, Myers RL, Devraj K, Coe CL, Lubach GR, Carruthers A et al (2015) A novel model for brain iron uptake: introducing the concept of regulation. J Cereb Blood Flow Metab 35:48–57
Torti FM, Torti SV (2002) Regulation of ferritin genes and protein. Blood 99:3505–3516
Truty J, Malpe R, Linder MC (2001) Iron prevents ferritin turnover in hepatic cells. J Biol Chem 276:48775–48780
Kleiger G, Mayor T (2014) Perilous journey: a tour of the ubiquitin-proteasome system. Trends Cell Biol 24:352–359
Sridhar S, Botbol Y, Macian F, Cuervo AM (2012) Autophagy and disease: always two sides to a problem. J Pathol 226:255–273
Feng Y, Yao Z, Klionsky DJ (2015) How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol 25:354–363
Stolz A, Ernst A, Dikic I (2014) Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 16:495–501
Park C, Cuervo AM (2013) Selective autophagy: talking with the UPS. Cell Biochem Biophys 67:3–13
Rudeck M, Volk T, Sitte N, Grune T (2000) Ferritin oxidation in vitro: implication of iron release and degradation by the 20S proteasome. IUBMB Life 49:451–456
Shringarpure R, Grune T, Mehlhase J, Davies KJ (2003) Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J Biol Chem 278:311–318
Kwok JC, Richardson DR (2004) Examination of the mechanism(s) involved in doxorubicin-mediated iron accumulation in ferritin: studies using metabolic inhibitors, protein synthesis inhibitors, and lysosomotropic agents. Mol Pharmacol 65:181–195
Mehlhase J, Sandig G, Pantopoulos K, Grune T (2005) Oxidation-induced ferritin turnover in microglial cells: role of proteasome. Free Radic Biol Med 38:276–285
De Domenico I, Ward DM, Kaplan J (2009) Specific iron chelators determine the route of ferritin degradation. Blood 114:4546–4551
De Domenico I, Vaughn MB, Li L, Bagley D, Musci G, Ward DM, Kaplan J (2006) Ferroportin-mediated mobilization of ferritin iron precedes ferritin degradation by the proteasome. EMBO J 25:5396–5404
Bulvik BE, Berenshtein E, Meyron-Holtz EG, Konijn AM, Chevion M (2012) Cardiac protection by preconditioning is generated via an iron-signal created by proteasomal degradation of iron proteins. PLoS One 7:e48947
Fedorko ME, Cross NL, Hirsch JG (1973) Appearance and distribution of ferritin in mouse peritoneal macrophages in vitro after uptake of heterologous erythrocytes. J Cell Biol 57:289–305
Marton PF (1975) Ultrastructural study of erythrophagocytosis in the rat bone marrow II. Iron metabolism in reticulum cells following red cell digestion. Scand J Haematol Suppl 23:27–48
Hernández-Yago J, Knecht E, Martínez-Ramón A, Grisolía S (1980) Autophagy of ferritin incorporated into the cytosol of Hela cells by liposomes. Cell Tissue Res 205:303–309
Hultcrantz R, Glaumann H (1987) Intracellular fate of ferritin in HeLa cells following microinjection. Exp Cell Res 171:203–212
Bridges KR, Hoffman KE (1986) The effects of ascorbic acid on the intracellular metabolism of iron and ferritin. J Biol Chem 261:14273–14277
Bridges KR (1987) Ascorbic acid inhibits lysosomal autophagy of ferritin. J Biol Chem 262:14773–14778
Larson JA, Howie HL, So M (2004) Neisseria meningitidis accelerates ferritin degradation in host epithelial cells to yield an essential iron source. Mol Microbiol 53:807–820
Xu P, Lin Y, Porter K, Liton PB (2014) Ascorbic acid modulation of iron homeostasis and lysosomal function in trabecular meshwork cells. J Ocul Pharmacol Ther 30:246–253
Ollinger K, Roberg K (1997) Nutrient deprivation of cultured rat hepatocytes increases the desferrioxamine-available iron pool and augments the sensitivity to hydrogen peroxide. J Biol Chem 272:23707–23711
Roberts S, Bomford A (1988) Ferritin iron kinetics and protein turnover in K562 cells. J Biol Chem 263:19181–19187
Sakaida I, Kyle ME, Farber JL (1990) Autophagic degradation of protein generates a pool of ferric iron required for the killing of cultured hepatocytes by an oxidative stress. Mol Pharmacol 37:435–442
Vaisman B, Fibach E, Konijn AM (1997) Utilization of intracellular ferritin iron for hemoglobin synthesis in developing human erythroid precursors. Blood 90:831–838
Goralska M, Nagar S, Fleisher LN, McGahan MC (2005) Differential degradation of ferritin H- and L-chains: accumulation of L-chain-rich ferritin in lens epithelial cells. Invest Ophthalmol Vis Sci 46:3521–3529
Kidane TZ, Sauble E, Linder MC (2006) Release of iron from ferritin requires lysosomal activity. Am J Physiol Cell Physiol 291:C445–455
Dong XP, Cheng X, Mills E, Delling M, Wang F, Kurz T, Xu H (2008) The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature 455:992–996
Kurz T, Eaton JW, Brunk UT (2011) The role of lysosomes in iron metabolism and recycling. Int J Biochem Cell Biol 43:1686–1697
Karlsson M, Frennesson C, Gustafsson T, Brunk UT, Nilsson SE, Kurz T (2013) Autophagy of iron-binding proteins may contribute to the oxidative stress resistance of ARPE-19 cells. Exp Eye Res 116:359–365
Zhang Y, Mikhael M, Xu D, Li Y, Soe-Lin S, Ning B, Li W, Nie G et al (2010) Lysosomal proteolysis is the primary degradation pathway for cytosolic ferritin and cytosolic ferritin degradation is necessary for iron exit. Antioxid Redox Signal 13:999–1009
Asano T, Komatsu M, Yamaguchi-Iwai Y, Ishikawa F, Mizushima N, Iwai K (2011) Distinct mechanisms of ferritin delivery to lysosomes in iron-depleted and iron-replete cells. Mol Cell Biol 31:2040–2052
Yang ND, Tan SH, Ng S, Shi Y, Zhou J, Tan KS, Wong WS, Shen HM (2014) Artesunate induces cell death in human cancer cells via enhancing lysosomal function and lysosomal degradation of ferritin. J Biol Chem 289:33425–33441
Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z et al (2014) Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol 16:1069–1079
Kishi-Itakura C, Koyama-Honda I, Itakura E, Mizushima N (2014) Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J Cell Sci 127:4089–4102
Kollara A, Brown TJ (2012) Expression and function of nuclear receptor co-activator 4: evidence of a potential role independent of co-activator activity. Cell Mol Life Sci 69:3895–3609
Dengjel J, Høyer-Hansen M, Nielsen MO, Eisenberg T, Harder LM, Schandorff S, Farkas T, Kirkegaard T et al (2012) Identification of autophagosome-associated proteins and regulators by quantitative proteomic analysis and genetic screens. Mol Cell Proteomics 11:M111.014035
Sharifi N, Hurt EM, Thomas SB, Farrar WL (2008) Effects of manganese superoxide dismutase silencing on androgen receptor function and gene regulation: implications for castration-resistant prostate cancer. Clin Cancer Res 14:6073–6080
Mitchell SH, Zhu W, Young CY (1999) Resveratrol inhibits the expression and function of the androgen receptor in LNCaP prostate cancer cells. Cancer Res 59:5892–5895
Gammella E, Buratti P, Cairo G, Recalcati S (2014) Macrophages: central regulators of iron balance. Metallomics 6:1336–1345
Roy CN, Mak HH, Akpan I, Losyev G, Zurakowski D, Andrews NC (2007) Hepcidin antimicrobial peptide transgenic mice exhibit features of the anemia of inflammation. Blood 109:4038–4044
Kroot JJ, Tjalsma H, Fleming RE, Swinkels DW (2011) Hepcidin in human iron disorders: diagnostic implications. Clin Chem 57:1650–1669
Finberg KE, Heeney MM, Campagna DR, Aydinok Y, Pearson HA, Hartman KR, Mayo MM, Samuel SM et al (2008) Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat Genet 40:569–571
Kautz L, Jung G, Nemeth E, Ganz T (2014) Erythroferrone contributes to recovery from anemia of inflammation. Blood 124:2569–2574
Weber GJ, Choe SE, Dooley KA, Paffett-Lugassy NN, Zhou Y, Zon LI (2005) Mutant-specific gene programs in the zebrafish. Blood 106:521–530
Goh SH, Josleyn M, Lee YT, Danner RL, Gherman RB, Cam MC, Miller JL (2007) The human reticulocyte transcriptome. Physiol Genomics 30:172–178
Merkerova M, Vasikova A, Bruchova H, Libalova H, Topinka J, Balascak I, Sram RJ, Brdicka R (2009) Differential gene expression in umbilical cord blood and maternal peripheral blood. Eur J Haematol 83:183–190
Peled M, Fisher EA (2014) Dynamic aspects of macrophage polarization during atherosclerosis progression and regression. Front Immunol 5:579
Hubler MJ, Peterson KR, Hasty AH (2015) Iron homeostasis: a new job for macrophages in adipose tissue? Trends Endocrinol Metab 26:101–109
Di Lorenzo D, Biasiotto G, Zanella I (2014) Source of iron overload in multiple sclerosis. Cell Mol Life Sci 71:3187–3189
Lunova M, Goehring C, Kuscuoglu D, Mueller K, Chen Y, Walther P, Deschemin JC, Vaulont S et al (2014) Hepcidin knockout mice fed with iron-rich diet develop chronic liver injury and liver fibrosis due to lysosomal iron overload. J Hepatol 61:633–641
Belsham DD, Evangelou A, Roy D, Duc VL, Brown TJ (1998) Regulation of gonadotropin-releasing hormone (GnRH) gene expression by 5 alpha-dihydrotestosterone in GnRH-secreting GT1-7 hypothalamic neurons. Endocrinology 139:1108–1114
Alen P, Claessens F, Schoenmakers E, Swinnen JV, Verhoeven G, Rombauts W, Peeters B (1999) Interaction of the putative androgen receptor-specific co-activator ARA70/ELE1alpha with multiple steroid receptors and identification of an internally deleted ELE1beta isoform. Mol Endocrinol 13:117–128
Siriett V, Nicholas G, Berry C, Watson T, Hennebry A, Thomas M, Ling N, Sharma M et al (2006) Myostatin negatively regulates the expression of the steroid receptor co-factor ARA70. J Cell Physiol 206:255–263
Kollara A, Brown TJ (2010) Variable expression of nuclear receptor co-activator 4 (NcoA4) during mouse embryonic development. J Histochem Cytochem 58:595–609
Damme M, Suntio T, Saftig P, Eskelinen EL (2015) Autophagy in neuronal cells: general principles and physiological and pathological functions. Acta Neuropathol 129:337–362
Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C et al (2010) Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A 107:14164–14169
Nixon RA, Yang DS (2012) Autophagy and neuronal cell death in neurological disorders. Cold Spring Harb Perspect Biol 4 pii: a008839
Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K et al (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889
Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M et al (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884
Nixon RA, Yang DS (2011) Autophagy failure in Alzheimer’s disease—locating the primary defect. Neurobiol Dis 43:38–45
Peric A, Annaert W (2015) Early etiology of Alzheimer’s disease: tipping the balance toward autophagy or endosomal dysfunction? Acta Neuropathol 129:363–381
Cai Z, Zhao B, Li K, Zhang L, Li C, Quazi SH, Tan Y (2012) Mammalian target of rapamycin: a valid therapeutic target through the autophagy pathway for Alzheimer’s disease? J Neurosci Res 90:1105–1118
Pasternak SH, Bagshaw RD, Guiral M, Zhang S, Ackerley CA, Pak BJ, Callahan JW, Mahuran DJ (2003) Presenilin-1, nicastrin, amyloid precursor protein, and gamma-secretase activity are co-localized in the lysosomal membrane. J Biol Chem 278:26687–26694
Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M et al (2010) Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141:1146–1158
Coffey EE, Beckel JM, Laties AM, Mitchell CH (2014) Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer’s disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience 263:111–124
Yang DS, Stavrides P, Mohan PS, Kaushik S, Kumar A, Ohno M, Schmidt SD, Wesson D et al (2011) Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer’s disease ameliorates amyloid pathologies and memory deficits. Brain 134:258–277
Caccamo A, Magrì A, Medina DX, Wisely EV, López-Aranda MF, Silva AJ, Oddo S (2013) mTOR regulates tau phosphorylation and degradation: implications for Alzheimer’s disease and other tauopathies. Aging Cell 12:370–380
Caccamo A, De Pinto V, Messina A, Branca C, Oddo S (2014) Genetic reduction of mammalian target of rapamycin ameliorates Alzheimer’s disease-like cognitive and pathological deficits by restoring hippocampal gene expression signature. J Neurosci 34:7988–7998
Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, Gonzales E, Burchett JM et al (2014) Enhancing astrocytic lysosome biogenesis facilitates Aβ clearance and attenuates amyloid plaque pathogenesis. J Neurosci 34:9607–9620
Polito VA, Li H, Martini-Stoica H, Wang B, Yang L, Xu Y, Swartzlander DB, Palmieri M et al (2014) Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol Med 6:1142–1160
Duce JA, Tsatsanis A, Cater MA, James SA, Robb E, Wikhe K, Leong SL, Perez K et al (2010) Iron-export ferroxidase activity of β-amyloid precursor protein is inhibited by zinc in Alzheimer’s disease. Cell 142:857–867
Lei P, Ayton S, Finkelstein DI, Spoerri L, Ciccotosto GD, Wright DK, Wong BX, Adlard PA et al (2012) Tau deficiency induces parkinsonism with dementia by impairing APP-mediated iron export. Nat Med 18:291–295
Silvestri L, Camaschella C (2008) A potential pathogenetic role of iron in Alzheimer’s disease. J Cell Mol Med 12:1548–1550
Hwang EM, Kim SK, Sohn JH, Lee JY, Kim Y, Kim YS, Mook-Jung I (2006) Furin is an endogenous regulator of alpha-secretase associated APP processing. Biochem Biophys Res Commun 349:654–659
Li X, Liu Y, Zheng Q, Yao G, Cheng P, Bu G, Xu H, Zhang YW (2013) Ferritin light chain interacts with PEN-2 and affects γ-secretase activity. Neurosci Lett 548:90–94
Ali-Rahmani F, Schengrund CL, Connor JR (2014) HFE gene variants, iron, and lipids: a novel connection in Alzheimer’s disease. Front Pharmacol 5:165
Bonda DJ, Liu G, Men P, Perry G, Smith MA, Zhu X (2012) Nanoparticle delivery of transition-metal chelators to the brain: Oxidative stress will never see it coming! CNS. Neurol Disord Drug Targets 11:81–85
Bartzokis G, Tishler TA (2000) MRI evaluation of basal ganglia ferritin iron and neurotoxicity in Alzheimer’s and Huntingon’s disease. Cell Mol Biol (Noisy-le-grand) 46:821–833
Raven EP, Lu PH, Tishler TA, Heydari P, Bartzokis G (2013) Increased iron levels and decreased tissue integrity in hippocampus of Alzheimer’s disease detected in vivo with magnetic resonance imaging. J Alzheimers Dis 37(1):127–36
Lopes KO, Sparks DL, Streit WJ (2008) Microglial dystrophy in the aged and Alzheimer’s disease brain is associated with ferritin immunoreactivity. Glia 56:1048–1060
Quintana C, Gutiérrez L (2010) Could a dysfunction of ferritin be a determinant factor in the aetiology of some neurodegenerative diseases? Biochim Biophys Acta 1800:770–782
Galazka-Friedman J, Bauminger ER, Szlachta K, Friedman A (2012) The role of iron in neurodegeneration—Mössbauer spectroscopy, electron microscopy, enzyme-linked immunosorbent assay and neuroimaging studies. J Phys Condens Matter 24:244106
Cox D, Carver JA, Ecroyd H (2014) Preventing α-synuclein aggregation: the role of the small heat-shock molecular chaperone proteins. Biochim Biophys Acta 1842:1830–1843
McNaught KS, Belizaire R, Jenner P, Olanow CW, Isacson O (2002) Selective loss of 20S proteasome alpha-subunits in the substantia nigra pars compacta in Parkinson’s disease. Neurosci Lett 326:155–158
Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D (2004) Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305:1292–1295
Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM (2006) Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A 103:5805–5810
Alvarez-Erviti L, Rodriguez-Oroz MC, Cooper JM, Caballero C, Ferrer I, Obeso JA, Schapira AH (2010) Chaperone-mediated autophagy markers in Parkinson disease brains. Arch Neurol 67:1464–1472
Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, Lichtenberg M, Menzies FM et al (2010) α-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J Cell Biol 190:1023–1037
Lynch-Day MA, Mao K, Wang K, Zhao M, Klionsky DJ (2012) The role of autophagy in Parkinson’s disease. Cold Spring Harb Perspect Med 2:a009357
Plun-Favreau H, Klupsch K, Moisoi N, Gandhi S, Kjaer S, Frith D, Harvey K, Deas E et al (2007) The mitochondrial protease HtrA2 is regulated by Parkinson’s disease-associated kinase PINK1. Nat Cell Biol 9:1243–1252
Gómez-Suaga P, Fdez E, Fernández B, Martínez-Salvador M, Blanca Ramírez M, Madero-Pérez J, Rivero-Ríos P, Fuentes JM et al (2014) Novel insights into the neurobiology underlying LRRK2-linked Parkinson’s disease. Neuropharmacology 85:45–56
MacLeod DA, Rhinn H, Kuwahara T, Zolin A, Di Paolo G, McCabe BD, MacCabe BD, Marder KS et al (2013) RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson’s disease risk. Neuron 77:425–439
van Veen S, Sørensen DM, Holemans T, Holen HW, Palmgren MG, Vangheluwe P (2014) Cellular function and pathological role of ATP13A2 and related P-type transport ATPases in Parkinson’s disease and other neurological disorders. Front Mol Neurosci 7:48
Murphy KE, Gysbers AM, Abbott SK, Tayebi N, Kim WS, Sidransky E, Cooper A, Garner B et al (2014) Reduced glucocerebrosidase is associated with increased α-synuclein in sporadic Parkinson’s disease. Brain 137:834–848
McGlinchey RP, Lee JC (2013) Emerging insights into the mechanistic link between α-synuclein and glucocerebrosidase in Parkinson’s disease. Biochem Soc Trans 41:1509–1512
McKeon JE, Sha D, Li L, Chin LS (2015) Parkin-mediated K63-polyubiquitination targets ubiquitin C-terminal hydrolase L1 for degradation by the autophagy-lysosome system. Cell Mol Life Sci 72:1811–1824
Kabuta T, Furuta A, Aoki S, Furuta K, Wada K (2008) Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem 283:23731–23738
Zavodszky E, Seaman MN, Moreau K, Jimenez-Sanchez M, Breusegem SY, Harbour ME, Rubinsztein DC (2014) Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat Commun 5:3828
Ivatt RM, Sanchez-Martinez A, Godena VK, Brown S, Ziviani E, Whitworth AJ (2014) Genome-wide RNAi screen identifies the Parkinson disease GWAS risk locus SREBF1 as a regulator of mitophagy. Proc Natl Acad Sci U S A 111:8494–8499
Cole NB, Murphy DD, Grider T, Rueter S, Brasaemle D, Nussbaum RL (2002) Lipid droplet binding and oligomerization properties of the Parkinson’s disease protein alpha-synuclein. J Biol Chem 277:6344–6352
Jiang P, Gan M, Lin WL, Yen SH (2014) Nutrient deprivation induces α-synuclein aggregation through endoplasmic reticulum stress response and SREBP2 pathway. Front Aging Neurosci 6:268
Settembre C, Ballabio A (2014) Lysosome: regulator of lipid degradation pathways. Trends Cell Biol 24:743–750
Savolainen MH, Richie CT, Harvey BK, Männistö PT, Maguire-Zeiss KA, Myöhänen TT (2014) The beneficial effect of a prolyl oligopeptidase inhibitor, KYP-2047, on alpha-synuclein clearance and autophagy in A30P transgenic mouse. Neurobiol Dis 68:1–15
Jang W, Kim HJ, Li H, Jo KD, Lee MK, Song SH, Yang HO (2014) 1,25-Dyhydroxyvitamin D3 attenuates rotenone-induced neurotoxicity in SH-SY5Y cells through induction of autophagy. Biochem Biophys Res Commun 451:142–147
Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Björklund A (2013) TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc Natl Acad Sci U S A 110:E1817–1826
Sian-Hülsmann J, Mandel S, Youdim MB, Riederer P (2011) The relevance of iron in the pathogenesis of Parkinson’s disease. J Neurochem 118:939–957
Weinreb O, Mandel S, Youdim MB, Amit T (2013) Targeting dysregulation of brain iron homeostasis in Parkinson’s disease by iron chelators. Free Radic Biol Med 62:52–64
Salazar J, Mena N, Hunot S, Prigent A, Alvarez-Fischer D, Arredondo M, Duyckaerts C, Sazdovitch V et al (2008) Divalent metal transporter 1 (DMT1) contributes to neurodegeneration in animal models of Parkinson’s disease. Proc Natl Acad Sci U S A 105:18578–18583
Jiang H, Song N, Xu H, Zhang S, Wang J, Xie J (2010) Up-regulation of divalent metal transporter 1 in 6-hydroxydopamine intoxication is IRE/IRP dependent. Cell Res 20:345–356
Jia W, Xu H, Du X, Jiang H, Xie J (2015) Ndfip1 attenuated 6-OHDA-induced iron accumulation via regulating the degradation of DMT1. Neurobiol Aging 36:1183–1193
Tabuchi M, Yanatori I, Kawai Y, Kishi F (2010) Retromer-mediated direct sorting is required for proper endosomal recycling of the mammalian iron transporter DMT1. J Cell Sci 123:756–766
Song N, Wang J, Jiang H, Xie J (2010) Ferroportin 1 but not hephaestin contributes to iron accumulation in a cell model of Parkinson’s disease. Free Radic Biol Med 48:332–341
Ayton S, Lei P, Duce JA, Wong BX, Sedjahtera A, Adlard PA, Bush AI, Finkelstein DI (2013) Ceruloplasmin dysfunction and therapeutic potential for Parkinson disease. Ann Neurol 73:554–559
Kostka M, Högen T, Danzer KM, Levin J, Habeck M, Wirth A, Wagner R, Glabe CG et al (2008) Single particle characterization of iron-induced pore-forming alpha-synuclein oligomers. J Biol Chem 283:10992–11003
He Q, Song N, Jia F, Xu H, Yu X, Xie J, Jiang H (2013) Role of α-synuclein aggregation and the nuclear factor E2-related factor 2/heme oxygenase-1 pathway in iron-induced neurotoxicity. Int J Biochem Cell Biol 45:1019–1030
Davies P, Moualla D, Brown DR (2011) Alpha-synuclein is a cellular ferrireductase. PLoS One 6, e15814
Allen GF, Toth R, James J, Ganley IG (2013) Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep 14:1127–1135
Friedman A, Galazka-Friedman J (2012) The history of the research of iron in parkinsonian substantia nigra. J Neural Transm 119:1507–1510
Tribl F, Asan E, Arzberger T, Tatschner T, Langenfeld E, Meyer HE, Bringmann G, Riederer P et al (2009) Identification of L-ferritin in neuromelanin granules of the human substantia nigra: a targeted proteomics approach. Mol Cell Proteomics 8:1832–1838
Paisán-Ruiz C, Li A, Schneider SA, Holton JL, Johnson R, Kidd D, Chataway J, Bhatia KP et al (2012) Widespread Lewy body and tau accumulation in childhood and adult onset dystonia-parkinsonism cases with PLA2G6 mutations. Neurobiol Aging 33:814–823
Popławska-Domaszewicz K, Florczak-Wyspiańska J, Kozubski W (2014) Update on neurodegeneration with brain iron accumulation. Neurol Neurochir Pol 48:206–213
Rinaldi DE, Corradi GR, Cuesta LM, Adamo HP, de Tezanos PF (2015) The Parkinson-associated human P5B-ATPase ATP13A2 protects against the iron-induced cytotoxicity. Biochim Biophys Acta 1850:1646–1655
Martin DD, Ladha S, Ehrnhoefer DE, Hayden MR (2015) Autophagy in Huntington disease and huntingtin in autophagy. Trends Neurosci 38:26–35
Koga H, Martinez-Vicente M, Arias E, Kaushik S, Sulzer D, Cuervo AM (2011) Constitutive upregulation of chaperone-mediated autophagy in Huntington’s disease. J Neurosci 31:18492–18505
Qi L, Zhang XD, Wu JC, Lin F, Wang J, DiFiglia M, Qin ZH (2012) The role of chaperone-mediated autophagy in huntingtin degradation. PLoS One 7, e46834
Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E et al (2010) Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci 13:567–576
Pal A, Severin F, Lommer B, Shevchenko A, Zerial M (2006) Huntingtin-HAP40 complex is a novel Rab5 effector that regulates early endosome motility and is up-regulated in Huntington’s disease. J Cell Biol 172:605–618
del Toro D, Alberch J, Lázaro-Diéguez F, Martín-Ibáñez R, Xifró X, Egea G, Canals JM (2009) Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol Biol Cell 20:1478–1492
Wong YC, Holzbaur EL (2014) The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci 34:1293–1305
Ochaba J, Lukacsovich T, Csikos G, Zheng S, Margulis J, Salazar L, Mao K, Lau AL et al (2014) Potential function for the huntingtin protein as a scaffold for selective autophagy. Proc Natl Acad Sci U S A 111:16889–16894
Metzger S, Saukko M, Van Che H, Tong L, Puder Y, Riess O, Nguyen HP (2010) Age at onset in Huntington’s disease is modified by the autophagy pathway: implication of the V471A polymorphism in Atg7. Hum Genet 128:453–459
Proenca CC, Stoehr N, Bernhard M, Seger S, Genoud C, Roscic A, Paganetti P, Liu S et al (2013) Atg4b-dependent autophagic flux alleviates Huntington’s disease progression. PLoS One 8, e68357
Sarkar S, Rubinsztein DC (2008) Huntington’s disease: degradation of mutant huntingtin by autophagy. FEBS J 275:4263–4270
Tsvetkov AS, Miller J, Arrasate M, Wong JS, Pleiss MA, Finkbeiner S (2010) A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc Natl Acad Sci U S A 107:16982–16987
Fernandez-Estevez MA, Casarejos MJ, López Sendon J, Garcia Caldentey J, Ruiz C, Gomez A, Perucho J, de Yebenes JG et al (2014) Trehalose reverses cell malfunction in fibroblasts from normal and Huntington’s disease patients caused by proteosome inhibition. PLoS One 9, e90202
Lai AY, Lan CP, Hasan S, Brown ME, McLaurin J (2014) scyllo-Inositol promotes robust mutant huntingtin protein degradation. J Biol Chem 289:3666–3676
Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, Lazarowski ER, Damian VA et al (2012) PGC-1α rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med 4:142ra197
Muller M, Leavitt BR (2014) Iron dysregulation in Huntington’s disease. J Neurochem 130:328–350
van den Bogaard SJ, Dumas EM, Roos RA (2013) The role of iron imaging in Huntington’s disease. Int Rev Neurobiol 110:241–250
Sánchez-Castañeda C, Squitieri F, Di Paola M, Dayan M, Petrollini M, Sabatini U (2015) The role of iron in gray matter degeneration in Huntington’s disease: a magnetic resonance imaging study. Hum Brain Mapp 36:50–66
Simmons DA, Casale M, Alcon B, Pham N, Narayan N, Lynch G (2007) Ferritin accumulation in dystrophic microglia is an early event in the development of Huntington’s disease. Glia 55:1074–1084
Dexter DT, Carayon A, Javoy-Agid F, Agid Y, Wells FR, Daniel SE, Lees AJ, Jenner P et al (1991) Alterations in the levels of iron, ferritin and other trace metals in Parkinson’s disease and other neurodegenerative diseases affecting the basal ganglia. Brain 114(Pt 4):1953–1975
Chen J, Marks E, Lai B, Zhang Z, Duce JA, Lam LQ, Volitakis I, Bush AI et al (2013) Iron accumulates in Huntington’s disease neurons: protection by deferoxamine. PLoS One 8, e77023
Lumsden AL, Henshall TL, Dayan S, Lardelli MT, Richards RI (2007) Huntingtin-deficient zebrafish exhibit defects in iron utilization and development. Hum Mol Genet 16:1905–1920
Hilditch-Maguire P, Trettel F, Passani LA, Auerbach A, Persichetti F, MacDonald ME (2000) Huntingtin: an iron-regulated protein essential for normal nuclear and perinuclear organelles. Hum Mol Genet 9:2789–2797
Nguyen T, Hamby A, Massa SM (2005) Clioquinol down-regulates mutant huntingtin expression in vitro and mitigates pathology in a Huntington’s disease mouse model. Proc Natl Acad Sci U S A 102:11840–11845
Cardoso F (2014) Differential diagnosis of Huntington’s disease: what the clinician should know. Neurodegener Dis Manag 4:67–72
Irwin DJ, Cairns NJ, Grossman M, McMillan CT, Lee EB, Van Deerlin VM, Lee VM, Trojanowski JQ (2015) Frontotemporal lobar degeneration: defining phenotypic diversity through personalized medicine. Acta Neuropathol 129:469–491
Van Langenhove T, van der Zee J, Van Broeckhoven C (2012) The molecular basis of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum. Ann Med 44:817–828
Thomas M, Alegre-Abarrategui J, Wade-Martins R (2013) RNA dysfunction and aggrephagy at the centre of an amyotrophic lateral sclerosis/frontotemporal dementia disease continuum. Brain 136:1345–1360
Bendotti C, Marino M, Cheroni C, Fontana E, Crippa V, Poletti A, De Biasi S (2012) Dysfunction of constitutive and inducible ubiquitin-proteasome system in amyotrophic lateral sclerosis: implication for protein aggregation and immune response. Prog Neurobiol 97:101–126
Galbiati M, Crippa V, Rusmini P, Cristofani R, Cicardi ME, Giorgetti E, Onesto E, Messi E et al (2014) ALS-related misfolded protein management in motor neurons and muscle cells. Neurochem Int 79:70–78
Bandyopadhyay U, Nagy M, Fenton WA, Horwich AL (2014) Absence of lipofuscin in motor neurons of SOD1-linked ALS mice. Proc Natl Acad Sci U S A 111:11055–11060
An T, Shi P, Duan W, Zhang S, Yuan P, Li Z, Wu D, Xu Z et al (2014) Oxidative stress and autophagic alteration in brainstem of SOD1-G93A mouse model of ALS. Mol Neurobiol 49:1435–1448
Crippa V, Boncoraglio A, Galbiati M, Aggarwal T, Rusmini P, Giorgetti E, Cristofani R, Carra S et al (2013) Differential autophagy power in the spinal cord and muscle of transgenic ALS mice. Front Cell Neurosci 7:234
Tian F, Morimoto N, Liu W, Ohta Y, Deguchi K, Miyazaki K, Abe K (2011) In vivo optical imaging of motor neuron autophagy in a mouse model of amyotrophic lateral sclerosis. Autophagy 7:985–992
Crippa V, Sau D, Rusmini P, Boncoraglio A, Onesto E, Bolzoni E, Galbiati M, Fontana E et al (2010) The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS). Hum Mol Genet 19:3440–3456
Kim J, Kim TY, Cho KS, Kim HN, Koh JY (2013) Autophagy activation and neuroprotection by progesterone in the G93A-SOD1 transgenic mouse model of amyotrophic lateral sclerosis. Neurobiol Dis 59:80–85
Zhang K, Shi P, An T, Wang Q, Wang J, Li Z, Duan W, Li C et al (2013) Food restriction-induced autophagy modulates degradation of mutant SOD1 in an amyotrophic lateral sclerosis mouse model. Brain Res 1519:112–119
Castillo K, Nassif M, Valenzuela V, Rojas F, Matus S, Mercado G, Court FA, van Zundert B et al (2013) Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy 9:1308–1320
Zhang X, Li L, Chen S, Yang D, Wang Y, Wang Z, Le W (2011) Rapamycin treatment augments motor neuron degeneration in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Autophagy 7:412–425
Otomo A, Kunita R, Suzuki-Utsunomiya K, Ikeda JE, Hadano S (2011) Defective relocalization of ALS2/alsin missense mutants to Rac1-induced macropinosomes accounts for loss of their cellular function and leads to disturbed amphisome formation. FEBS Lett 585:730–736
Moustaqim-Barrette A, Lin YQ, Pradhan S, Neely GG, Bellen HJ, Tsuda H (2014) The amyotrophic lateral sclerosis 8 protein, VAP, is required for ER protein quality control. Hum Mol Genet 23:1975–1989
Paul P, de Belleroche J (2014) The role of D-serine and glycine as co-agonists of NMDA receptors in motor neuron degeneration and amyotrophic lateral sclerosis (ALS). Front Synaptic Neurosci 6:10
Bentmann E, Haass C, Dormann D (2013) Stress granules in neurodegeneration—lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. FEBS J 280:4348–4370
Buchan JR, Kolaitis RM, Taylor JP, Parker R (2013) Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153:1461–1474
Ryu HH, Jun MH, Min KJ, Jang DJ, Lee YS, Kim HK, Lee JA (2014) Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol Aging 35:2822–2831
Figley MD, Bieri G, Kolaitis RM, Taylor JP, Gitler AD (2014) Profilin 1 associates with stress granules and ALS-linked mutations alter stress granule dynamics. J Neurosci 34:8083–8097
Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, Krobitsch S (2007) Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell 18:1385–1396
Korac J, Schaeffer V, Kovacevic I, Clement AM, Jungblut B, Behl C, Terzic J, Dikic I (2013) Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J Cell Sci 126:580–592
Osaka M, Ito D, Yagi T, Nihei Y, Suzuki N (2015) Evidence of a link between ubiquilin 2 and optineurin in amyotrophic lateral sclerosis. Hum Mol Genet 24:1617–1629
Wong YC, Holzbaur EL (2014) Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A 111:E4439–4448
Pottier C, Bieniek KF, Finch N, van de Vorst M, Baker M, Perkersen R, Brown P, Ravenscroft T et al (2015) Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol 130:77–92
Rusten TE, Simonsen A (2008) ESCRT functions in autophagy and associated disease. Cell Cycle 7:1166–1172
Isaacs AM, Johannsen P, Holm I, Nielsen JE, consortium F (2011) Frontotemporal dementia caused by CHMP2B mutations. Curr Alzheimer Res 8:246–251
Vollrath JT, Sechi A, Dreser A, Katona I, Wiemuth D, Vervoorts J, Dohmen M, Chandrasekar A et al (2014) Loss of function of the ALS protein SigR1 leads to ER pathology associated with defective autophagy and lipid raft disturbances. Cell Death Dis 5, e1290
Wu Q, Liu M, Huang C, Liu X, Huang B, Li N, Zhou H, Xia XG (2015) Pathogenic Ubqln2 gains toxic properties to induce neuron death. Acta Neuropathol 129:417–428
Rea SL, Majcher V, Searle MS, Layfield R (2014) SQSTM1 mutations—bridging Paget disease of bone and ALS/FTLD. Exp Cell Res 325:27–37
Lattante S, de Calbiac H, Le Ber I, Brice A, Ciura S, Kabashi E (2015) Sqstm1 knock-down causes a locomotor phenotype ameliorated by rapamycin in a zebrafish model of ALS/FTLD. Hum Mol Genet 24:1682–1690
Nalbandian A, Donkervoort S, Dec E, Badadani M, Katheria V, Rana P, Nguyen C, Mukherjee J et al (2011) The multiple faces of valosin-containing protein-associated diseases: inclusion body myopathy with Paget’s disease of bone, frontotemporal dementia, and amyotrophic lateral sclerosis. J Mol Neurosci 45:522–531
Yamamoto A, Simonsen A (2011) The elimination of accumulated and aggregated proteins: a role for aggrephagy in neurodegeneration. Neurobiol Dis 43:17–28
Walker AK, Soo KY, Sundaramoorthy V, Parakh S, Ma Y, Farg MA, Wallace RH, Crouch PJ et al (2013) ALS-associated TDP-43 induces endoplasmic reticulum stress, which drives cytoplasmic TDP-43 accumulation and stress granule formation. PLoS One 8, e81170
Wang IF, Guo BS, Liu YC, Wu CC, Yang CH, Tsai KJ, Shen CK (2012) Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc Natl Acad Sci U S A 109:15024–15029
Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, Ando DM, Tsvetkov A, Pleiss M et al (2014) Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol 10:677–685
Bose JK, Huang CC, Shen CK (2011) Regulation of autophagy by neuropathological protein TDP-43. J Biol Chem 286:44441–44448
DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA et al (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–256
Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ, Consortium I (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72:257–268
Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC et al (2014) C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507:195–200
Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ (2013) The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics 29:499–503
Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, Halloran MA, Gleeson PA et al (2014) C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet 23:3579–3595
Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–646
Tao Z, Wang H, Xia Q, Li K, Jiang X, Xu G, Wang G, Ying Z (2015) Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet 24:2426–2441
Tanaka Y, Chambers JK, Matsuwaki T, Yamanouchi K, Nishihara M (2014) Possible involvement of lysosomal dysfunction in pathological changes of the brain in aged progranulin-deficient mice. Acta Neuropathol Commun 2:78
Ahmed Z, Sheng H, Xu YF, Lin WL, Innes AE, Gass J, Yu X, Wuertzer CA et al (2010) Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am J Pathol 177:311–324
Smith KR, Damiano J, Franceschetti S, Carpenter S, Canafoglia L, Morbin M, Rossi G, Pareyson D et al (2012) Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet 90:1102–1107
Götzl JK, Mori K, Damme M, Fellerer K, Tahirovic S, Kleinberger G, Janssens J, van der Zee J et al (2014) Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol 127:845–860
Stagi M, Klein ZA, Gould TJ, Bewersdorf J, Strittmatter SM (2014) Lysosome size, motility and stress response regulated by fronto-temporal dementia modifier TMEM106B. Mol Cell Neurosci 61:226–240
Jiang T, Yu JT, Zhu XC, Zhang QQ, Cao L, Wang HF, Tan MS, Gao Q et al (2014) Temsirolimus attenuates tauopathy in vitro and in vivo by targeting tau hyperphosphorylation and autophagic clearance. Neuropharmacology 85:121–130
Ferrari R, Hernandez DG, Nalls MA, Rohrer JD, Ramasamy A, Kwok JB, Dobson-Stone C, Brooks WS et al (2014) Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol 13:686–699
Oba H, Araki T, Ohtomo K, Monzawa S, Uchiyama G, Koizumi K, Nogata Y, Kachi K et al (1993) Amyotrophic lateral sclerosis: T2 shortening in motor cortex at MR imaging. Radiology 189:843–846
Langkammer C, Enzinger C, Quasthoff S, Grafenauer P, Soellinger M, Fazekas F, Ropele S (2010) Mapping of iron deposition in conjunction with assessment of nerve fiber tract integrity in amyotrophic lateral sclerosis. J Magn Reson Imaging 31:1339–1345
Kwan JY, Jeong SY, Van Gelderen P, Deng HX, Quezado MM, Danielian LE, Butman JA, Chen L et al (2012) Iron accumulation in deep cortical layers accounts for MRI signal abnormalities in ALS: correlating 7 tesla MRI and pathology. PLoS One 7, e35241
Ignjatović A, Stević Z, Lavrnić S, Daković M, Bačić G (2013) Brain iron MRI: a biomarker for amyotrophic lateral sclerosis. J Magn Reson Imaging 38:1472–1479
Yu J, Qi F, Wang N, Gao P, Dai S, Lu Y, Su Q, Du Y et al (2014) Increased iron level in motor cortex of amyotrophic lateral sclerosis patients: an in vivo MR study. Amyotroph Lateral Scler Frontotemporal Degener 15:357–361
Adachi Y, Sato N, Saito Y, Kimura Y, Nakata Y, Ito K, Kamiya K, Matsuda H et al (2015) Usefulness of SWI for the detection of iron in the motor cortex in amyotrophic lateral sclerosis. J Neuroimaging 25:443–451
Kasarskis EJ, Tandon L, Lovell MA, Ehmann WD (1995) Aluminum, calcium, and iron in the spinal cord of patients with sporadic amyotrophic lateral sclerosis using laser microprobe mass spectroscopy: a preliminary study. J Neurol Sci 130:203–208
Offen D, Barhum Y, Melamed E, Embacher N, Schindler C, Ransmayr G (2009) Spinal cord mRNA profile in patients with ALS: comparison with transgenic mice expressing the human SOD-1 mutant. J Mol Neurosci 38:85–93
Olsen MK, Roberds SL, Ellerbrock BR, Fleck TJ, McKinley DK, Gurney ME (2001) Disease mechanisms revealed by transcription profiling in SOD1-G93A transgenic mouse spinal cord. Ann Neurol 50:730–740
Jeong SY, Rathore KI, Schulz K, Ponka P, Arosio P, David S (2009) Dysregulation of iron homeostasis in the CNS contributes to disease progression in a mouse model of amyotrophic lateral sclerosis. J Neurosci 29:610–619
Winkler EA, Sengillo JD, Sagare AP, Zhao Z, Ma Q, Zuniga E, Wang Y, Zhong Z et al (2014) Blood-spinal cord barrier disruption contributes to early motor-neuron degeneration in ALS-model mice. Proc Natl Acad Sci U S A 111:E1035–1042
Bamm VV, Harauz G (2014) Hemoglobin as a source of iron overload in multiple sclerosis: does multiple sclerosis share risk factors with vascular disorders? Cell Mol Life Sci 71:1789–1798
Mitchell RM, Simmons Z, Beard JL, Stephens HE, Connor JR (2010) Plasma biomarkers associated with ALS and their relationship to iron homeostasis. Muscle Nerve 42:95–103
Ikeda K, Hirayama T, Takazawa T, Kawabe K, Iwasaki Y (2012) Relationships between disease progression and serum levels of lipid, urate, creatinine and ferritin in Japanese patients with amyotrophic lateral sclerosis: a cross-sectional study. Intern Med 51:1501–1508
Nadjar Y, Gordon P, Corcia P, Bensimon G, Pieroni L, Meininger V, Salachas F (2012) Elevated serum ferritin is associated with reduced survival in amyotrophic lateral sclerosis. PLoS One 7, e45034
Veyrat-Durebex C, Corcia P, Mucha A, Benzimra S, Mallet C, Gendrot C, Moreau C, Devos D et al (2014) Iron metabolism disturbance in a French cohort of ALS patients. Biomed Res Int 2014:485723
Hozumi I, Hasegawa T, Honda A, Ozawa K, Hayashi Y, Hashimoto K, Yamada M, Koumura A et al (2011) Patterns of levels of biological metals in CSF differ among neurodegenerative diseases. J Neurol Sci 303:95–99
Conti A, Iannaccone S, Sferrazza B, De Monte L, Cappa S, Franciotta D, Olivieri S, Alessio M (2008) Differential expression of ceruloplasmin isoforms in the cerebrospinal fluid of amyotrophic lateral sclerosis patients. Proteomics Clin Appl 2:1628–1637
Nandar W, Connor JR (2011) HFE gene variants affect iron in the brain. J Nutr 141:729S–739S
Liu Y, Lee SY, Neely E, Nandar W, Moyo M, Simmons Z, Connor JR (2011) Mutant HFE H63D protein is associated with prolonged endoplasmic reticulum stress and increased neuronal vulnerability. J Biol Chem 286:13161–13170
Su XW, Lee SY, Mitchell RM, Stephens HE, Simmons Z, Connor JR (2013) H63D HFE polymorphisms are associated with increased disease duration and decreased muscle superoxide dismutase-1 expression in amyotrophic lateral sclerosis patients. Muscle Nerve 48:242–246
Nandar W, Neely EB, Simmons Z, Connor JR (2014) H63D HFE genotype accelerates disease progression in animal models of amyotrophic lateral sclerosis. Biochim Biophys Acta 1842:2413–2426
Deschauer M, Gaul C, Behrmann C, Prokisch H, Zierz S, Haack TB (2012) C19orf12 mutations in neurodegeneration with brain iron accumulation mimicking juvenile amyotrophic lateral sclerosis. J Neurol 259:2434–2439
Santillo AF, Skoglund L, Lindau M, Eeg-Olofsson KE, Tovi M, Engler H, Brundin RM, Ingvast S et al (2009) Frontotemporal dementia-amyotrophic lateral sclerosis complex is simulated by neurodegeneration with brain iron accumulation. Alzheimer Dis Assoc Disord 23:298–300
Schweitzer K, Decker E, Zhu L, Miller RE, Mirra SS, Spina S, Ghetti B, Wang M et al (2006) Aberrantly regulated proteins in frontotemporal dementia. Biochem Biophys Res Commun 348:465–472
De Reuck JL, Deramecourt V, Auger F, Durieux N, Cordonnier C, Devos D, Defebvre L, Moreau C et al (2014) Iron deposits in post-mortem brains of patients with neurodegenerative and cerebrovascular diseases: a semi-quantitative 7.0 T magnetic resonance imaging study. Eur J Neurol 21:1026–1031
Gazzina S, Premi E, Zanella I, Biasiotto G, Archetti S, Cosseddu M, Scarpini E, Galimberti D, Serpente M, Gasparotti R, Padovani A, Borroni B. Iron in frontotemporal lobar degeneration: a new subcortical pathological pathway? (accepted for publication in Neurodegener Dis)
Dusek P, Jankovic J, Le W (2012) Iron dysregulation in movement disorders. Neurobiol Dis 46:1–18
Arber CE, Li A, Houlden H, Wray S (2015) Insights into molecular mechanisms of disease in neurodegeneration with brain iron accumulation: unifying theories. Neuropathol Appl Neurobiol. doi:10.1111/nan.12242
Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, Sugai K, Kasai-Yoshida E, Sawaura N et al (2013) De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet 45(445–449):449e441
Zanella I, Derosas M, Corrado M, Cocco E, Cavadini P, Biasiotto G, Poli M, Verardi R et al (2008) The effects of frataxin silencing in HeLa cells are rescued by the expression of human mitochondrial ferritin. Biochim Biophys Acta 1782:90–98
Martelli A, Puccio H (2014) Dysregulation of cellular iron metabolism in Friedreich ataxia: from primary iron-sulfur cluster deficit to mitochondrial iron accumulation. Front Pharmacol 5:130
Simon D, Seznec H, Gansmuller A, Carelle N, Weber P, Metzger D, Rustin P, Koenig M et al (2004) Friedreich ataxia mouse models with progressive cerebellar and sensory ataxia reveal autophagic neurodegeneration in dorsal root ganglia. J Neurosci 24:1987–1995
Deguchi K, Ikeda K, Kume K, Takata T, Kokudo Y, Kamada M, Touge T, Honjo N et al (2015) Significance of the hot-cross bun sign on T2*-weighted MRI for the diagnosis of multiple system atrophy. J Neurol. doi:10.1007/s00415-015-7728-1
Sugiyama A, Ito S, Suichi T, Sakurai T, Mukai H, Yokota H, Yonezu T, Kuwabara S (2015) Putaminal hypointensity on T2*-weighted MR imaging is the most practically useful sign in diagnosing multiple system atrophy: a preliminary study. J Neurol Sci 349:174–178
Matsusue E, Fujii S, Kanasaki Y, Kaminou T, Ohama E, Ogawa T (2009) Cerebellar lesions in multiple system atrophy: postmortem MR imaging-pathologic correlations. AJNR Am J Neuroradiol 30:1725–1730
Visanji NP, Collingwood JF, Finnegan ME, Tandon A, House E, Hazrati LN (2013) Iron deficiency in parkinsonism: region-specific iron dysregulation in Parkinson’s disease and multiple system atrophy. J Parkinsons Dis 3:523–537
Benarroch EE, Schmeichel AM, Parisi JE, Low PA (2015) Putative neuropathological interactions in MSA: focus in the rostral ventrolateral medulla. Clin Auton Res 25:77–80
Tanji K, Odagiri S, Maruyama A, Mori F, Kakita A, Takahashi H, Wakabayashi K (2013) Alteration of autophagosomal proteins in the brain of multiple system atrophy. Neurobiol Dis 49:190–198
Schwarz L, Goldbaum O, Bergmann M, Probst-Cousin S, Richter-Landsberg C (2012) Involvement of macroautophagy in multiple system atrophy and protein aggregate formation in oligodendrocytes. J Mol Neurosci 47:256–266
Lee JH, Han YH, Kang BM, Mun CW, Lee SJ, Baik SK (2013) Quantitative assessment of subcortical atrophy and iron content in progressive supranuclear palsy and parkinsonian variant of multiple system atrophy. J Neurol 260:2094–2101
Han YH, Lee JH, Kang BM, Mun CW, Baik SK, Shin YI, Park KH (2013) Topographical differences of brain iron deposition between progressive supranuclear palsy and parkinsonian variant multiple system atrophy. J Neurol Sci 325:29–35
Boelmans K, Holst B, Hackius M, Finsterbusch J, Gerloff C, Fiehler J, Münchau A (2012) Brain iron deposition fingerprints in Parkinson’s disease and progressive supranuclear palsy. Mov Disord 27:421–427
Ebrahim AS, Kulathingal J, Murray ME, Casey-Castanedes M, Dickson DW, Yen SH, Sevlever D (2011) A proteomic study identifies different levels of light chain ferritin in corticobasal degeneration and progressive supranuclear palsy. Acta Neuropathol 122:727–736
Bras JM (2013) Lysosomal storage disorders and iron. Int Rev Neurobiol 110:251–275
Jeyakumar M, Williams I, Smith D, Cox TM, Platt FM (2009) Critical role of iron in the pathogenesis of the murine gangliosidoses. Neurobiol Dis 34:406–416
Cuervo AM, Dice JF (2000) Age-related decline in chaperone-mediated autophagy. J Biol Chem 275:31505–31513
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Biasiotto, G., Di Lorenzo, D., Archetti, S. et al. Iron and Neurodegeneration: Is Ferritinophagy the Link?. Mol Neurobiol 53, 5542–5574 (2016). https://doi.org/10.1007/s12035-015-9473-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-015-9473-y