Abstract
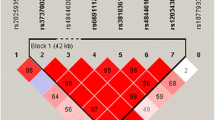
Rare variants of phospholipase D3 (PLD3) have been identified as Alzheimer’s disease (AD) susceptibility loci, whereas little is known about the potential role of common variants in the progression of AD. To examine the impact of genetic variations in PLD3 on neuroimaging phenotypes in a large non-demented population. A total of 261 normal cognition (NC) and 456 mild cognitive impairment (MCI) individuals from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database are included in our analysis. Multiple linear regression models were applied to examine the association between four single-nucleotide polymorphisms (SNPs; rs7249146, rs4490097, rs12151243, and rs10407447) with the florbetapir retention on florbetapir 18F amyloid positron emission tomography (AV45-PET), the cerebral metabolic rate for glucose (CMRgl) on 18F-fluorodeoxyglucose PET (FDG-PET), and regional volume on magnetic resonance imaging (MRI) at baseline and in the cohort study. We did not detect any significant associations of PLD3 SNPs with florbetapir retention on AV45-PET. In the analysis of FDG-PET, rs10407447 was associated with the CMRgl in the left angular gyrus and bilateral posterior cingulate cortex in the MCI group. Regarding the MRI analysis, rs10407447 was also associated with bilateral inferior lateral ventricle and lateral ventricle volume in MCI group. The main findings of our study provide evidence that support the possible role of PLD3 common variants in influencing AD-related neuroimaging phenotypes. Nevertheless, further work is necessary to explain the functional mechanisms of differences and confirm the causal variants.





Similar content being viewed by others
References
Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL (2006) Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 63(2):168–174. doi:10.1001/archpsyc.63.2.168
Jiang T, Yu JT, Tian Y, Tan L (2013) Epidemiology and etiology of Alzheimer’s disease: from genetic to non-genetic factors. Curr Alzheimer Res 10(8):852–867
Jiang T, Yu JT, Tan L (2012) Novel disease-modifying therapies for Alzheimer’s disease. J Alzheimer’s Dis : JAD 31(3):475–492. doi:10.3233/JAD-2012-120640
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V et al (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 41(10):1088–1093. doi:10.1038/ng.440
Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D et al (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 41(10):1094–1099. doi:10.1038/ng.439
Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF et al (1993) Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology 43(8):1467–1472
Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, Harari O, Norton J et al (2014) Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature 505(7484):550–554. doi:10.1038/nature12825
Mattay VS, Goldberg TE, Sambataro F, Weinberger DR (2008) Neurobiology of cognitive aging: insights from imaging genetics. Biol Psychol 79(1):9–22. doi:10.1016/j.biopsycho.2008.03.015
Salmon DP, Lange KL (2001) Cognitive screening and neuropsychological assessment in early Alzheimer’s disease. Clin Geriatr Med 17(2):229–254
Bennett DA, Schneider JA, Arvanitakis Z, Kelly JF, Aggarwal NT, Shah RC, Wilson RS (2006) Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology 66(12):1837–1844. doi:10.1212/01.wnl.0000219668.47116.e6
Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, Risacher SL, Nho K et al (2010) Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement : J Alzheimers Assoc 6(3):265–273. doi:10.1016/j.jalz.2010.03.013
Landau SM, Harvey D, Madison CM, Koeppe RA, Reiman EM, Foster NL, Weiner MW, Jagust WJ (2011) Associations between cognitive, functional, and FDG-PET measures of decline in AD and MCI. Neurobiol Aging 32(7):1207–1218. doi:10.1016/j.neurobiolaging.2009.07.002
Wang J, Yu JT, Tan L (2014) PLD3 in Alzheimer’s Disease. Mol Neurobiol. doi:10.1007/s12035-014-8779-5
Clark CM, Schneider JA, Bedell BJ, Beach TG, Bilker WB, Mintun MA, Pontecorvo MJ, Hefti F et al (2011) Use of florbetapir-PET for imaging beta-amyloid pathology. JAMA 305(3):275–283. doi:10.1001/jama.2010.2008
Small GW, Mazziotta JC, Collins MT, Baxter LR, Phelps ME, Mandelkern MA, Kaplan A, La Rue A et al (1995) Apolipoprotein E type 4 allele and cerebral glucose metabolism in relatives at risk for familial Alzheimer disease. JAMA 273(12):942–947
Jagust WJ, Landau SM (2012) Apolipoprotein E, not fibrillar beta-amyloid, reduces cerebral glucose metabolism in normal aging. J Neurosci : Off J Soc Neurosci 32(50):18227–18233. doi:10.1523/JNEUROSCI.3266-12.2012
Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J (2004) Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer’s dementia. Proc Natl Acad Sci U S A 101(1):284–289. doi:10.1073/pnas.2635903100
Minoshima S, Giordani B, Berent S, Frey KA, Foster NL, Kuhl DE (1997) Metabolic reduction in the posterior cingulate cortex in very early Alzheimer’s disease. Ann Neurol 42(1):85–94. doi:10.1002/ana.410420114
Cattaneo Z, Silvanto J, Pascual-Leone A, Battelli L (2009) The role of the angular gyrus in the modulation of visuospatial attention by the mental number line. NeuroImage 44(2):563–568. doi:10.1016/j.neuroimage.2008.09.003
Johnson SC, Christian BT, Okonkwo OC, Oh JM, Harding S, Xu G, Hillmer AT, Wooten DW et al (2014) Amyloid burden and neural function in people at risk for Alzheimer’s disease. Neurobiol Aging 35(3):576–584. doi:10.1016/j.neurobiolaging.2013.09.028
Wu TC, Wilde EA, Bigler ED, Yallampalli R, McCauley SR, Troyanskaya M, Chu Z, Li X et al (2010) Evaluating the relationship between memory functioning and cingulum bundles in acute mild traumatic brain injury using diffusion tensor imaging. J Neurotrauma 27(2):303–307. doi:10.1089/neu.2009.1110
Chou YY, Lepore N, de Zubicaray GI, Carmichael OT, Becker JT, Toga AW, Thompson PM (2008) Automated ventricular mapping with multi-atlas fluid image alignment reveals genetic effects in Alzheimer’s disease. NeuroImage 40(2):615–630. doi:10.1016/j.neuroimage.2007.11.047
Nestor SM, Rupsingh R, Borrie M, Smith M, Accomazzi V, Wells JL, Fogarty J, Bartha R (2008) Ventricular enlargement as a possible measure of Alzheimer’s disease progression validated using the Alzheimer’s disease neuroimaging initiative database. Brain : J Neurol 131(Pt 9):2443–2454. doi:10.1093/brain/awn146
Ridha BH, Barnes J, Bartlett JW, Godbolt A, Pepple T, Rossor MN, Fox NC (2006) Tracking atrophy progression in familial Alzheimer’s disease: a serial MRI study. Lancet Neurol 5(10):828–834. doi:10.1016/S1474-4422(06)70550-6
Qiu A, Fennema-Notestine C, Dale AM, Miller MI (2009) Regional shape abnormalities in mild cognitive impairment and Alzheimer’s disease. NeuroImage 45(3):656–661
Pedersen KM, Finsen B, Celis JE, Jensen NA (1998) Expression of a novel murine phospholipase D homolog coincides with late neuronal development in the forebrain. J Biol Chem 273(47):31494–31504
Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS et al (2003) Dynamics of gray matter loss in Alzheimer’s disease. J Neurosci : Off J Soc Neurosci 23(3):994–1005
Kremen WS, Prom-Wormley E, Panizzon MS, Eyler LT, Fischl B, Neale MC, Franz CE, Lyons MJ et al (2010) Genetic and environmental influences on the size of specific brain regions in midlife: the VETSA MRI study. NeuroImage 49(2):1213–1223. doi:10.1016/j.neuroimage.2009.09.043
Madsen SK, Ho AJ, Hua X, Saharan PS, Toga AW, Jack CR Jr, Weiner MW, Thompson PM (2010) 3D maps localize caudate nucleus atrophy in 400 Alzheimer’s disease, mild cognitive impairment, and healthy elderly subjects. Neurobiol Aging 31(8):1312–1325. doi:10.1016/j.neurobiolaging.2010.05.002
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). This research was also supported by National Natural Science Foundation of China (81171209, 81371406, 81000544), the Shandong Provincial Outstanding Medical Academic Professional Program, and the Qingdao Key Health Discipline Development Fund.
Conflict of interest
The authors declare that they have no competing interests
Author information
Authors and Affiliations
Consortia
Corresponding authors
Additional information
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 1184 kb)
Rights and permissions
About this article
Cite this article
Wang, C., Wang, HF., Tan, MS. et al. Impact of Common Variations in PLD3 on Neuroimaging Phenotypes in Non-demented Elders. Mol Neurobiol 53, 4343–4351 (2016). https://doi.org/10.1007/s12035-015-9370-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-015-9370-4