Abstract
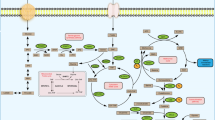
Breast cancer is a prevalent malignant tumor, posing a significant threat to women's health globally due to its increasing incidence and tendency to affect younger patients. Protein tyrosine phosphatases (PTPs) are a class of enzymes that have emerged as potential targets for various tumors, including breast cancer, because they can modulate oncogenic tyrosine kinases, which are both tumor-suppressive and oncogenic. The regulation of tyrosine phosphorylation levels is crucial for cell proliferation and differentiation. Although the clinical biomarker potential of PTPs is not fully explored, there is evidence to suggest that they may serve as clinical biomarkers and therapeutic targets for breast cancer. We found that increased expression levels of PTPN11 and PTPN3 were associated with a higher risk of death in patients with breast cancer, while PTPN11 and PTPN18 are significantly associated with overall survival in patients with estrogen receptor-positive (ER+) breast cancer. Meanwhile, PTPN11 expression was found to be negatively associated with survival in patients with ER+ breast cancer. Furthermore, PTPN11 exposes a metabolic vulnerability to breast cancer metastasis via dysregulated ceramide metabolism. Therefore, we speculate that PTPN11 has the potential to serve as a therapeutic target for breast cancer by regulating lipid metabolism reprogramming.










Similar content being viewed by others
Data availability
The datasets analyzed during the current study are available in the “TCGA” dataset” (http://ualcan.path.uab.edu/cgi-bin/TCGAExHeatMap2.pl?size=25&cancer=BRCA).
References
Huang J, et al. Global incidence and mortality of breast cancer: a trend analysis. Aging (Albany NY). 2021;13:5748–803. https://doi.org/10.18632/aging.202502.
Liang Y, Zhang H, Song X, Yang Q. Metastatic heterogeneity of breast cancer: molecular mechanism and potential therapeutic targets. Semin Cancer Biol. 2020;60:14–27. https://doi.org/10.1016/j.semcancer.2019.08.012.
Eugenio DS, et al. Breast cancer features in women under the age of 40 years. Rev Assoc Med Bras. 2016;1992(62):755–61. https://doi.org/10.1590/1806-9282.62.08.755.
Cardoso F, et al. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-updagger. Ann Oncol. 2019;30:1194–220. https://doi.org/10.1093/annonc/mdz173.
Tang W, et al. Digital breast tomosynthesis plus mammography, magnetic resonance imaging plus mammography and mammography alone: a comparison of diagnostic performance in symptomatic women. Clin Hemorheol Microcirc. 2017;66:105–16. https://doi.org/10.3233/CH-16242.
Nunes-Xavier CE, Martin-Perez J, Elson A, Pulido R. Protein tyrosine phosphatases as novel targets in breast cancer therapy. Biochim Biophys Acta. 1836;211–226:2013. https://doi.org/10.1016/j.bbcan.2013.06.001.
Kim M, Baek M, Kim DJ. Protein tyrosine signaling and its potential therapeutic implications in carcinogenesis. Curr Pharm Des. 2017;23:4226–46. https://doi.org/10.2174/1381612823666170616082125.
Dubreuil V, Sap J, Harroch S. Protein tyrosine phosphatase regulation of stem and progenitor cell biology. Semin Cell Dev Biol. 2015;37:82–9. https://doi.org/10.1016/j.semcdb.2014.09.012.
Wang S, et al. UCSCXenaShiny: an R/CRAN package for interactive analysis of UCSC Xena data. Bioinformatics. 2022;38:527–9. https://doi.org/10.1093/bioinformatics/btab561.
Chandrashekar DS, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–58. https://doi.org/10.1016/j.neo.2017.05.002.
Tang Z, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–102. https://doi.org/10.1093/nar/gkx247.
Goel MK, Khanna P, Kishore J. Understanding survival analysis: Kaplan-Meier estimate. Int J Ayurveda Res. 2010;1:274–8. https://doi.org/10.4103/0974-7788.76794.
Modhukur V, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10:277–88. https://doi.org/10.2217/epi-2017-0118.
Zhang C, et al. SurvivalMeth: a web server to investigate the effect of DNA methylation-related functional elements on prognosis. Brief Bioinform. 2021. https://doi.org/10.1093/bib/bbaa162.
Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013. https://doi.org/10.1126/scisignal.2004088.
Szklarczyk D, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–12. https://doi.org/10.1093/nar/gkaa1074.
Warde-Farley D, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214-220. https://doi.org/10.1093/nar/gkq537.
Dennis G Jr, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3.
Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. https://doi.org/10.1038/s41467-019-09234-6.
Han H, et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380–6. https://doi.org/10.1093/nar/gkx1013.
Li T, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–14. https://doi.org/10.1093/nar/gkaa407.
Gray KA, Seal RL, Tweedie S, Wright MW, Bruford EA. A review of the new HGNC gene family resource. Hum Genomics. 2016;10:6. https://doi.org/10.1186/s40246-016-0062-6.
Ubhi T, Brown GW. Exploiting DNA replication stress for cancer treatment. Cancer Res. 2019;79:1730–9. https://doi.org/10.1158/0008-5472.CAN-18-3631.
Tang M, O’Grady S, Crown J, Duffy MJ. MYC as a therapeutic target for the treatment of triple-negative breast cancer: preclinical investigations with the novel MYC inhibitor, MYCi975. Breast Cancer Res Treat. 2022;195:105–15. https://doi.org/10.1007/s10549-022-06673-6.
Peng D, Fu M, Wang M, Wei Y, Wei X. Targeting TGF-beta signal transduction for fibrosis and cancer therapy. Mol Cancer. 2022;21:104. https://doi.org/10.1186/s12943-022-01569-x.
Miricescu D, et al. PI3K/AKT/mTOR signaling pathway in breast cancer: from molecular landscape to clinical aspects. Int J Mol Sci. 2020. https://doi.org/10.3390/ijms22010173.
Chen J, Zhao X, Yuan Y, Jing JJ. The expression patterns and the diagnostic/prognostic roles of PTPN family members in digestive tract cancers. Cancer Cell Int. 2020;20:238. https://doi.org/10.1186/s12935-020-01315-7.
Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–46. https://doi.org/10.1038/nrm2039.
Sahu R, Pattanayak SP. Strategic developments & future perspective on gene therapy for breast cancer: role of mTOR and Brk/ PTK6 as molecular targets. Curr Gene Ther. 2020;20:237–58. https://doi.org/10.2174/1566523220999200731002408.
Akdeniz D, et al. Effects of chemotherapy on contralateral breast cancer risk in BRCA1 and BRCA2 mutation carriers: a nationwide cohort study. Breast. 2022;61:98–107. https://doi.org/10.1016/j.breast.2021.12.007.
Li Y, et al. Long non-coding RNA UCA1 promotes breast cancer by upregulating PTP1B expression via inhibiting miR-206. Cancer Cell Int. 2019;19:275. https://doi.org/10.1186/s12935-019-0958-z.
Yu M, et al. PTP1B markedly promotes breast cancer progression and is regulated by miR-193a-3p. FEBS J. 2019;286:1136–53. https://doi.org/10.1111/febs.14724.
Geng Q, Xian R, Yu Y, Chen F, Li R. SHP-1 acts as a tumor suppressor by interacting with EGFR and predicts the prognosis of human breast cancer. Cancer Biol Med. 2021;19:468–85. https://doi.org/10.20892/j.issn.2095-3941.2020.0501.
Yu L, et al. HePTP promotes migration and invasion in triple-negative breast cancer cells via activation of Wnt/beta-catenin signaling. Biomed Pharmacother. 2019;118: 109361. https://doi.org/10.1016/j.biopha.2019.109361.
Shen J, et al. Role of DUSP1/MKP1 in tumorigenesis, tumor progression and therapy. Cancer Med. 2016;5:2061–8. https://doi.org/10.1002/cam4.772.
Li Z, Xu W, Ren X, Xu J, Chen J. Puerarin promotes DUSP1 expression by regulating miR-133a-3p in breast cancer. Mol Med Rep. 2019;19:205–12. https://doi.org/10.3892/mmr.2018.9682.
Glondu-Lassis M, et al. PTPL1/PTPN13 regulates breast cancer cell aggressiveness through direct inactivation of SRC kinase. Cancer Res. 2010;70:5116–26. https://doi.org/10.1158/0008-5472.CAN-09-4368.
Revillion F, et al. Expression of the putative tumor suppressor gene PTPN13/PTPL1 is an independent prognostic marker for overall survival in breast cancer. Int J Cancer. 2009;124:638–43. https://doi.org/10.1002/ijc.23989.
Carlucci A, et al. PTPD1 supports receptor stability and mitogenic signaling in bladder cancer cells. J Biol Chem. 2010;285:39260–70. https://doi.org/10.1074/jbc.M110.174706.
Jongmans MC, et al. Cancer risk in patients with Noonan syndrome carrying a PTPN11 mutation. Eur J Hum Genet. 2011;19:870–4. https://doi.org/10.1038/ejhg.2011.37.
Bentires-Alj M, et al. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64:8816–20. https://doi.org/10.1158/0008-5472.CAN-04-1923.
Wang LB, et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. 2021;39:509-528 e520. https://doi.org/10.1016/j.ccell.2021.01.006.
Zhang J, Zhang F, Niu R. Functions of Shp2 in cancer. J Cell Mol Med. 2015;19:2075–83. https://doi.org/10.1111/jcmm.12618.
Yuan Y, et al. SHP2 promotes proliferation of breast cancer cells through regulating Cyclin D1 stability via the PI3K/AKT/GSK3beta signaling pathway. Cancer Biol Med. 2020;17:707–25. https://doi.org/10.20892/j.issn.2095-3941.2020.0056.
Wang HM, et al. The catalytic region and PEST domain of PTPN18 distinctly regulate the HER2 phosphorylation and ubiquitination barcodes. Cell Res. 2014;24:1067–90. https://doi.org/10.1038/cr.2014.99.
Saddoughi SA, Ogretmen B. Diverse functions of ceramide in cancer cell death and proliferation. Adv Cancer Res. 2013;117:37–58. https://doi.org/10.1016/B978-0-12-394274-6.00002-9.
Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18:33–50. https://doi.org/10.1038/nrc.2017.96.
Acknowledgements
The authors thank the financial support of Southeast University. The abbreviations are presented in Table S4.
Funding
This work was financially supported by the Zhishan Scholars Programs of Southeast University (2242021R41070).
Author information
Authors and Affiliations
Contributions
Data curation and formal analysis, SQ, TW, and HW; original draft, HW.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
The studies involving human participants were reviewed and approved by Ningxia Medical University General Hospital Scientific Research Ethics Committee (KYLL-2023-0440).
Consent to participate
The patients/participants provided their written informed consent to participate in this study.
Consent for publication
Not applicable for that section.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Qiao, S., Wang, T. & Wang, H. Dysregulated ceramides metabolism via PTPN11 exposes a metabolic vulnerability to breast cancer metastasis. Med Oncol 40, 310 (2023). https://doi.org/10.1007/s12032-023-02187-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-023-02187-3