Abstract
Purpose of Review
Cardiovascular toxicity is a leading cause of mortality among cancer survivors and has become increasingly prevalent due to improved cancer survival rates. In this review, we synthesize evidence illustrating how common cancer therapeutic agents, such as anthracyclines, human epidermal growth factors receptors (HER2) monoclonal antibodies, and tyrosine kinase inhibitors (TKIs), have been evaluated in cardiomyocytes (CMs) derived from human-induced pluripotent stem cells (hiPSCs) to understand the underlying mechanisms of cardiovascular toxicity. We place this in the context of precision cardio-oncology, an emerging concept for personalizing the prevention and management of cardiovascular toxicities from cancer therapies, accounting for each individual patient’s unique factors. We outline steps that will need to be addressed by multidisciplinary teams of cardiologists and oncologists in partnership with regulators to implement future applications of hiPSCs in precision cardio-oncology.
Recent Findings
Current prevention of cardiovascular toxicity involves routine screenings and management of modifiable risk factors for cancer patients, as well as the initiation of cardioprotective medications. Despite recent advancements in precision cardio-oncology, knowledge gaps remain and limit our ability to appropriately predict with precision which patients will develop cardiovascular toxicity. Investigations using patient-specific CMs facilitate pharmacological discovery, mechanistic toxicity studies, and the identification of cardioprotective pathways. Studies with hiPSCs demonstrate that patients with comorbidities have more frequent adverse responses, compared to their counterparts without cardiac disease. Further studies utilizing hiPSC modeling should be considered, to evaluate the impact and mitigation of known cardiovascular risk factors, including blood pressure, body mass index (BMI), smoking status, diabetes, and physical activity in their role in cardiovascular toxicity after cancer therapy. Future real-world applications will depend on understanding the current use of hiPSC modeling in order for oncologists and cardiologists together to inform their potential to improve our clinical collaborative practice in cardio-oncology.
Summary
When applying such in vitro characterization, it is hypothesized that a safety score can be assigned to each individual to determine who has a greater probability of developing cardiovascular toxicity. Using hiPSCs to create personalized models and ultimately evaluate the cardiovascular toxicity of individuals’ treatments may one day lead to more patient-specific treatment plans in precision cardio-oncology while reducing cardiovascular disease (CVD) morbidity and mortality.
Similar content being viewed by others
Introduction
Cardiovascular adverse effects of cancer therapies are a leading cause of morbidity and mortality in cancer survivors and contribute to the world-wide burden of CVD [1]. Newer cancer therapies have resulted in increased survival rates, juxtaposed with an increase in cardiovascular complications, especially in those with CVD risk factors such as diabetes, hypertension, and hyperlipidemia [1]. Failure to predict and prevent CVD in cancer patients and survivors can therefore lead to detrimental long-term consequences of cardiovascular toxicity [2]. Thus, the field of cardio-oncology is dedicated to the prevention, screening, early diagnosis, and treatment of cardiovascular toxicity in the growing numbers of cancer survivors.
The prediction of cardiovascular toxicity for the goal of prevention includes identifying risk factors, comorbid conditions, and regular screening through echocardiography, electrocardiography (ECG), serum biomarkers, and other methods as applicable [3]. Predicting risk for toxicities has always been difficult, with the prediction of risk for targeted therapies proving especially challenging. There are no proven methods of precisely predicting which particular individuals are most likely to develop cardiovascular toxicity from cancer therapies. Drug classification, clinical, and host variables can provide some insight into who may be at risk, but alone, these methods are inadequate. The benefits of accurate risk prediction are obvious, e.g., guiding treatment decisions before the first dose is given. This can be particularly important for agents with a lengthy half-life and high potential to cause life-threatening cardiac complications, e.g., immune checkpoint inhibitors and their association with myocarditis. A personalized approach for each patient may help optimize efforts at prediction and prevention. Such an approach may integrate clinical factors, treatment plans, and individual characteristics such as those elucidated by genomics (assessing the individual’s entire complement of genes), proteomics (assessing the individual’s entire complement of proteins), and other “omics.” The integration of such a personalized medicine (or “precision medicine” approach) may present an opportunity to address these needs for better prediction and prevention in cardio-oncology. Therefore, to pinpoint individuals at risk and modify their care in response, precision medicine may prove especially beneficial. This integration may allow for novel insights into pathomechanisms and the development of more precise and cost-effective risk prediction tools, with the overall objective of better therapy decisions before, during, and after cancer treatment.
While various omics compose a large portion of precision medicine, hiPSCs provide a medium for studying omics on an individualized basis and are in their own right a method of individualizing the study of patient cardiotoxic responses, deserving particular attention in precision cardio-oncology. HiPSCs have the potential to revolutionize the care of patients by offering an individualized assessment of cardiovascular risk regarding short-term and long-term side effects of cancer therapies. HiPSCs can be used to offer in vitro screening which can potentially be used to develop and assign a precise and personal safety score that could be developed in the future to help oncologists and cardiologists anticipate and prevent cardiotoxicity. HiPSCs may thereby play a role in predictive and preventive personalized medicine. HiPSC-derived cardiomyocytes (hiPSC-CMs) are starting to play a critical role in cardio-oncology, specifically in disease modeling, researching new cancer therapies, and investigating the toxicity of current therapies [3]. The advantages of using hiPSCs over animal models are the elimination of interspecies variability and preventing many of the ethical or legal restrictions associated with investigations using human embryonic stem cells. Further, hiPSCs can be patient-specific, diminishing interindividual variability and biases, which can be particularly beneficial for population-based studies.
In this review, we evaluate the current data regarding the reprogramming of somatic cells into hiPSCs, differentiation strategies, and the use of hiPSCs in the study of three common pharmacologic cancer therapy drug classes: anthracyclines, HER2 monoclonal antibodies, and TKIs. We delve specifically into how hiPSC models can help elucidate the mechanisms of cardiovascular toxicity from these three drug classes, with the ultimate goal of preventing these toxicities. We ultimately describe current basic research and potential translation to future clinical practice in precision cardio-oncology.
Precision Cardio-Oncology
Currently cardio-oncology is limited by a lack of individualized prevention methods. However, precision medicine could advance the understanding of the risk of cardiovascular toxicity in each patient, with the eventual goal of reducing morbidity and mortality for all oncology patients. Precision cardio-oncology can allow for a patient-centered treatment approach for cardiovascular toxicity, aiming for a personalized cancer treatment regimen based on personalized tumor characteristics of each patient. Precision cardio-oncology may help us advance the understanding of the disease phenotype in cancer patients exposed to common oncology therapies. One way in which this can be accomplished is using hiPSCS to dissect how a patient’s cardiovascular risk, cancer type, and cancer treatments play a role in the potential development of cardiovascular toxicity. Research is also being pursued using hiPSCs to further understand the mechanisms underlying cardiovascular toxicity, in order to reduce the mortality and morbidity in cancer patients.
Transcriptomics, genomics, proteomics, metabolomics, microbiomics, and other omics, as well as pharmacokinetics and pharmacodynamics, of cancer therapy signaling pathways, receptors, and ligands all play a role in precision cardio-oncology. Incorporating data from different perspectives (e.g., proteomics, kinomics, genomics, and metabolomics) can offer mechanistic insight, uncover important risk factors, and may pinpoint new treatment options [4, 5]. A crucial question is whether biomarkers and other omic components that relate to drug efficacy and resistance also have the ability to predict cardiovascular toxicity in response to cancer therapy. This provides opportunities to apply precision medicine to increase the understanding, management, and prevention of cardiovascular toxicity caused by cancer therapies. Yet, the effect on clinical practice is yet to be determined. To consider the context in which precision cardio-oncology involving hiPSCs will ultimately be practiced, cardiovascular risk factors, risk stratification, potential for modeling individualized cardiotoxic patient responses to therapy, opportunities for prevention, and the future of using hiPSCs for prevention, prediction, and precision must be considered. Eventually, studies incorporating various omics in the setting of hiPSCs may become the standard for prediction cardiovascular toxicity in cardio-oncology.
Cardiovascular Risk Factors and Disparities
Cancer and CVD share many common risk factors, and it is believed that CVD and cancer can directly influence one another. The American Heart Association (AHA) recognizes blood pressure, BMI, smoking status, fasting glucose, diet, total cholesterol, and physical activity as risk factors in the development of both cardiovascular risk factors and cancer [6,7,8,9,10,11]. Furthermore, it is hypothesized that cardiometabolic risk worsens in patients with cancer even prior to treatment due to metabolic derangement [12, 13]. Additionally, treatment modalities for cancer, such as pharmacologic cancer therapy, radiation therapy, and less so surgery, can also lead to further worsening of cardiac disease [12,13,14,15,16,17].
Health disparities in cardiovascular toxicities from cancer therapies are often overlooked yet need to be addressed. It is important to consider populations with the highest cardio-oncologic risk, who may also be the most likely to benefit from predictive and preventive efforts. African Americans have higher incidence and mortality than their white counterparts in all combined malignancies and in cardiovascular toxicities from cancer therapies [18,19,20,21,22,23]. These disparities can be attributed to a complex interaction of differences in access to healthcare, life stressors, socioeconomic status, and comorbidities [24, 25]. These disparities should be considered when planning cardiovascular toxicity management, and future hiPSC modeling studies should include participants of all races to properly account for racial health disparities.
There is extensive evidence that the management of cardiac risk factors including hypertension, hyperlipidemia, and diabetes, as well a lifestyle changes such as diet and exercise, will decrease adverse cardiotoxic effects [26•, 27]. HiPSCs have opened a new avenue to investigate the effects of cancer therapies on the cardiovascular system. HiPSCs have been shown to be more representative of certain aspects of the individual patient than animal models such as in individuals with medical comorbidities, especially genetically determined comorbidities, which have been shown to respond differently than hiPSCs from healthy individuals. In particular, hiPSCs from patients with underlying long QT syndrome, dilated cardiomyopathy, and hypertrophic cardiomyopathy were shown to have adverse responses (with vulnerability to fatal arrhythmias) to cancer therapy, compared to individuals without cardiac comorbidities [28, 29]. Inclusion of patient-specific cardiac risk factors deduced by hiPSCs in the decision-making process in regard to pharmacologic cancer therapeutic agent administration and dosing is a glimpse into precision medicine with great potential for a positive impact on cardio-oncologic clinical practice.
Risk Stratification
Each individual responds differently to pharmacologic cancer therapeutic agents and has a different risk profile for cardiovascular toxicity. Estimates for cardiovascular toxicity from pharmacologic cancer therapeutic agents have a wide range: 8–26% for doxorubicin, 7–28% for trastuzumab, and 5–30% for paclitaxel [29]. There is an immense need to better risk stratify patients to mitigate toxicity. Current approaches begin with identifying risk factors and having regularly scheduled screening with echocardiography, EKGs, and biomarkers.
Multiple groups have attempted to create scores to predict risk of major cardiovascular events or toxicities within the cancer groups. A scoring system to predict major cardiovascular events after early-stage breast cancer uses age, prior heart failure, atrial fibrillation, peripheral vascular disease, hypertension, ischemic heart disease, diabetes, chronic kidney disease, chronic obstructive pulmonary disease (COPD), and cerebrovascular disease to risk stratify patients for cardiovascular toxicity at 5 years and 10 years [30•]. A purported scoring system to help risk stratify all patients undergoing chemotherapy uses patient and treatment factors to suggest monitoring and management recommendations [31].
HiPSCs have been utilized to create 3D cardiac models that profile the cardiotoxic effects of various cancer drugs [1]. Compared to animal models, hiPSC models have been found to be more representative of human cardiac physiology [32]. It is hypothesized that these in vitro studies could be used to help create a proposed safety score for each individual patient and inform propensity to develop cardiovascular toxicity. Currently, no validated model derived using hiPSCs exists that calculates the risk for cardiovascular toxicity following the initiation of pharmacologic cancer therapeutic agents. However, studies have already commenced utilizing hiPSC-CMs to help further risk stratify patients. In a small study, hiPSC-CMs from 11 healthy patients and 2 patients with kidney cancer were obtained and exposed to multiple TKIs to determine which had the most side effects in these patients [33]. This study found that sorafenib, regorafenib, and ponatinib were the most likely to induce side effects. Ponatinib was also found to inhibit pro-survival signaling pathways in hiPSC cardiomyocytes [34, 35].
Additionally, millions of SNPs across the genome were evaluated using genome-wide association studies (GWAS) [29, 36, 37]. However, it has been difficult to evaluate and validate the findings from the GWAS studies; often the validation process has relied on cardiac biopsies as the gold standard. It is believed that hiPSCs can play a major role in the validation process to confirm the GWAS studies [29]. Furthermore, when utilizing multiple molecular profiles to characterize hiPSC-derived lines, such as epigenomics, metabolomics, and transcriptomics, unique, patient-specific signatures are uncovered that could be more telling in regard to cardiotoxicity risk stratification. Modeling using hiPSCs may be the missing part of the puzzle in helping to create a complete and validated risk stratification system for cardiotoxicity in patients being treated with pharmacologic cancer therapies.
In order to develop and create a novel personalized safety score utilizing hiPSCs, further studies would be required to collect more information on risk factors for cardiovascular toxicity. These studies would demand robust collection of patient-specific hiPSCs from individuals with various risk profiles. Only after these studies are completed and findings analyzed can the information be used to stratify patients’ cardiovascular toxicity profiles into groups based on low or high toxicity. Deeper risk stratification will aid in our progress in the journey towards cardio-oncologic precision medicine. As the prediction of future cardiac events and risk scoring systems improve, so will the overall protection of cancer patients and survivors.
Modeling Using hiPSCs
Reprogramming Somatic Cells into hiPSCs
Researchers and clinicians are turning to hiPSCs for a broad range of patient-specific disease modeling. The hiPSCs are typically generated by acquiring primary human cells for reprogramming by key transcription factors, termed “Yamanaka” factors, Oct3/4, Sox2, Klf4, and c-Myc [38]. The Yamanaka factors induce the pluripotent cells to exhibit the states of lineage plasticity that are capable of differentiation into many cell types [38]. Oct3/4, Sox2, Nanog, and Lin28 (Thomson factors) were also described as being sufficient for reprogramming human cells [39]. Multiple reprogramming methods are in use today, the most common being Sendai retroviral transduction, with episomal iPSCs being used more often in recent years, eliminating mutations caused by viral integrations [40]. Other methods involve mRNAs, microRNA/mRNA transfection, minicircle vectors, and lentiviral transduction. Some of these methods are capable of reprogramming primary cells and generating transgene-free and virus-free iPSCs. Transcriptomic and epigenetic profiling of the resultant hiPSCs indicated that reprogramming of human primary fibroblasts did not differ significantly among the methods used, with standardized protocols [41].
Some primary somatic cell types used to derive iPSCs include fibroblasts, smooth muscle cells, skin cells (keratinocytes), urine cells, and peripheral blood mononuclear cells (PBMCs). The selected donor primary cell type is important since molecular profiles may be distinct and cell-type–specific. An example study involved hiPSCs derived from umbilical cord blood cells and neonatal keratinocytes, which had discrete patterns of epigenetic markers that aid in transcriptional regulation (i.e., DNA methylation) [42, 43]. Inconsistencies were seen in DNA methylation profiles upon differentiation to six different cell types representing all three germ layers. The differentiated cells retained some DNA methylation patterns of the parental cell types, although the functional implications of such residual epigenetic marks remain unclear [44••]. Consequently, many factors play a role in variations seen among hiPSC lines, such as primary human source cell type, viral load, passage number, and growth and sub-culturing techniques. The key molecular signatures or biomarkers retained during this process may be important for imminent modeling strategies. Time is also an issue as some primary cells require culturing and expansion before reprogramming.
Differentiation of hiPSCs into hiPSC-Derived Cardiomyocytes
The goal of hiPSC differentiation is to achieve a model system such as hiPSC-CMs to mimic the genetic, epigenetic, metabolic, biochemical, and physiological states of mature, adult cardiomyocytes (CMs), although they may be more comparable to immature CMs than adult CMs. The stages from hiPSC to CMs involve (1) epithelial to mesenchymal transition; (2) mesoderm induction; (3) mesoderm specification (towards cardiac); (4) cardiac specification (cardiac progenitor cells, CPCs); (5) cardiac lineage differentiation, which leads to the immature CMs; and (6) further development into mature CMs [45]. The likely range of time it takes to start seeing the initiation of beating is approximately 8–10 days [46]. There are numerous individual protocols for the differentiation of hiPSCs into CMs, and although most are analogous, there are still many researchers that are “fine-tuning” the protocols by further optimizing media, growth conditions, and nutrient supplementation, as well as creating more specific and detailed differentiation timelines. Reported by multiple groups, the effects of various methods of optimization for hiPSC-derived CMs specifically lead to increased cell mass and cellular expansion capabilities, improved contractility and force generation, enhanced calcium (Ca2+) kinetics and cycling, and increased membrane capacitance and activation or suppression of specific metabolic pathways, which eventually yield high numbers of mature hiPSC-CMs [47,48,49,50,51,52,53,54].
Differentiation of hiPSCs into Various Cardiac Cell Types
Aside from the heavily studied hiPSC-CMs, researchers are looking into the differentiation of multiple cardiac cell types that are derived from the CPC stage, such as cardiac mesenchymal stromal cells [55], epicardial cells [56], smooth muscle cells, endothelial cells, and cardiac fibroblasts and the myoblasts that develop from them which are known to spontaneously differentiate [57]. Some researchers are also defining hiPSC-CM subtypes such as atrial, ventricular, or nodal [58]. A research group imaged and characterized action potentials (APs) in subtype-specific hiPSC-CMs [58]. They used cardiac lineage-specific gene promoters to express a transmembrane voltage sensor linked to green fluorescent protein (GFP) and red fluorescent protein (RFP) which are brought into close proximity upon membrane depolarization, experiencing fluorescence resonance energy transfer (FRET), making GFP dimmer and RFP brighter. The ratio of RFP/GFP is equal to the AP, and these data closely match the accompanying gene expression data, verifying the method [58].
Co-culturing of multiple cardiac cell types may boost hiPSC-CM maturation due to juxtacrine and paracrine signaling. For instance, a few properties of enhanced maturation of hiPSC-CMs when co-culturing endothelial cells with CPCs were CM-specific gene and protein expression, size increase, upregulation of genes involved in sarcomere formation and electromechanical coupling, and development of T-tubule-like extensions. Co-cultures were characterized by determining the percentage of cells that had specific markers over the time course [59]. A myriad of researchers also use fluorescence-activated cell sorting (FACS) or flow cytometry to enumerate and separate cell types. Many functional assessments are in use to further characterize cells and cultures, including assays for shear stress, nitric oxide production, tube formation, vascular networks, and perfusion [60].
These approaches are becoming more in-depth and precise with the use of omics techniques like genomics (single-nucleotide polymorphisms, SNPs), transcriptomics (mRNA gene expression), epigenomics (DNA methylation, histone marks, chromatin conformation), or metabolomics (cellular/tissue metabolites, intermediates, and subsequent products). Such components of more complete molecular and phenotypic maps of human physiology and pathophysiology will likely enhance modeling of cardiotoxicity. In this setting, hiPSCs are also promising tools to model cardiotoxicity. To date, hiPSCs have primarily been used for retrospective studies, given the timeline needed for the process of deriving specific cells from hiPSCs while patients undergo cancer therapy. Various intricate details of differentiation protocols are important for cell maturity, production, and integrity, whether in culture or cryofrozen waiting to be cultured again when the patient may need them the most. In the following sections, we dive into three classes of pharmacologic cancer therapeutic agents and give examples of current investigations using hiPSCs in modeling cardiotoxicity.
Modeling Anthracyclines
Extracted from the Streptomyces bacterium, anthracyclines are used in various cancer treatment regimens. The most commonly used anthracyclines are doxorubicin, daunorubicin, epirubicin, and idarubicin. The mechanisms of action include the creation of free radicals, interference with macromolecular biosynthesis, DNA adduct formation, and interference with topoisomerase II activity [61]. Despite their effectiveness in cancer treatment, anthracyclines can cause cardiovascular toxicity that negatively affects patients’ outcomes and seriously limits their oncological therapeutic opportunities. The effect of anthracyclines on topoisomerase IIα (TOP2α) is generally considered the molecular basis of anthracycline activity as this isoform has the main chemotherapeutic role during replication and cell proliferation [62, 63]. In contrast, topoisomerase II beta (Top2β) has been associated with side effects of anthracyclines such as cardiovascular toxicity. It was shown that most hiPSC-CMs do not express TOP2α and that TOP2β was expressed in all CMs, which is also true for adult terminally differentiated CMs. TOP2β inhibition leads to the activation of protein kinase B, inhibition of adenosine monophosphate–activated protein kinase (AMPK), and more pronounced phosphorylation of further downstream elements such as glycogen synthase kinase 3β [64]. The resulting impaired energy signaling by AMPK inhibition likely initiates a negative feedback loop that potentiates doxorubicin-induced energy deficits and seriously compromises heart resistance to additional energy stress. The resulting cellular changes lead to the generation of harmful reactive oxygen species (ROS; i.e., superoxide, O2−; and hydrogen peroxide, H2O2), impaired Ca2+ homeostasis, mitochondrial damage, and induction of cell apoptosis [65]. ROS contribute to mitochondrial dysfunction by an effect on NADH dehydrogenase of the electron transport chain in the presence of molecular oxygen [66]. Topoisomerases are important enzymes required to create necessary chromatin remodeling to enable DNA replication and transcription. Topoisomerase II, which creates double-stranded breaks to enable this remodeling, is expressed as isoenzymes Top2α and Top2β. Top2α is expressed in actively dividing cells, while Top2β is present in quiescent cells such as CMs [66]. In CMs, anthracyclines bind Top2β, inducing mitochondrial dysfunction and activating β-adrenergic signaling, p53, and apoptosis [67]. In animal studies, the absence of Top2β has protected subjects against anthracycline-induced cardiovascular toxicity [68, 69].
Since anthracycline-induced cardiovascular toxicity can cause long-lasting effects, it is imperative to appropriately predict and identify cardiotoxic drug–patient combinations. This endeavor is difficult due to the inability to accurately represent human heart tissue for drug safety screening methodologies [70]. HiPSCs have become a novel tool to mimic the attributes of CMs and effectively test their reactions to various drug candidates (e.g., anthracyclines) to predict cardiovascular toxicity [70] (Table 1). In current clinical practice, the prediction of anthracycline-induced cardiovascular toxicity remains imprecise. The use of hiPSC-CMs could help reveal a patient’s susceptibility to cardiovascular toxicity at the level of a single cell [72]. The investigators described a powerful approach that uses CMs from patient-specific hiPSCs to investigate the differential sensitivity of individuals with breast cancer to doxorubicin-induced cardiomyopathy. They used hiPSCs from individuals with breast cancer who were treated with doxorubicin and either did or did not develop doxorubicin-induced cardiomyopathy. To study doxorubicin cardiovascular toxicity, the authors used current state-of-the art methods to generate high-purity preparations of CMs from the hiPSCs. Remarkably, in an extensive array of in vitro assays, the hiPSC-CMs from individuals who experienced doxorubicin-induced cardiovascular toxicity reproducibly showed greater sensitivity to doxorubicin toxicity than did hiPSC-CMs from treated patients who did not exhibit toxicity. In the same study, hiPSC-CMs from breast cancer patients who suffered doxorubicin-induced cardiovascular toxicity were used as a tool to understand the genetic indicators and molecular mechanisms that made them more sensitive to further cardiovascular toxicity [72]. Such information could be used for subsequent prediction and prevention if determined in vitro prior to in vivo therapy. Although this novel technique is not a definitive predictor of patient-specific cardiovascular toxicity, it can be used as a foundation to understand the cell-specific characteristics that cause predilection for cardiovascular toxicity, for more personalization of therapeutics in the future [72] (Table 1).
Transcriptome-wide analysis in hiPSC-CMs and mouse cardiac cells exposed to doxorubicin identified that the RNA-binding protein (RBP), Quaking (QKI), was significantly downregulated. Lentiviral plasmid or adeno-associated virus 9 (AAV9)–mediated overexpression and knockout experiments of the QKI gene, Qki5, in multiple mouse cell types and lines lead to the conclusion that QKI regulated specific circular RNAs (circRNAs) which demonstrated anti-apoptotic effects and mediated doxorubicin-induced cardiovascular toxicity [78]. The future will continue to yield more effector molecules and potential therapeutic candidates. Another group described an engineered, multi-chamber tissue chip complete with a 3D human micro-organ system consisting of colon cancer tumor spheroids in a fibrin gel, hiPSC-CM spheroids in a separate compartment in fibrin gel, and hiPSC-ECs in a microfluidic channel compartment in between. The chip was used to evaluate cardiotoxicity within 48 h of doxorubicin administration to gauge the drug effects on beat frequency and conduction velocity of hiPSC-CMs, as measured by electrodes within the chip and intracellular Ca2+ concentration. The group observed that doxorubicin reduced both the beating rate and conduction velocity at or near the half inhibitory concentration (IC50; drug efficacy), consistent with previous in vivo literature [79]. GWAS indicated that a variant in the retinoic acid receptor gamma gene, RARG, was associated with doxorubicin-induced cardiotoxicity, but follow-up replication and functional analyses were lacking. Other investigators accumulated case (left ventricular ejection fraction (LVEF) < 55%) and control patients with and without doxorubicin-induced cardiotoxicity. Patient-specific hiPSC-CMs from cases retained that sensitivity in vitro, whereas the wild-type (WT) hiPSC-CMs remained insensitive. When the mutation of RARG-S427L was genetically corrected in cases, hiPSC-CMs had a reduced cardiotoxic effects upon doxorubicin exposure, whereas the doxorubicin-induced cardiotoxicity increased when the RARG gene was knocked out using genetic editing technologies in controls [80]. These studies highlight how patient-derived hiPSCs can be used to infer direct or causal roles in the toxic effects of a common chemotherapy agent. Using hiPSCs in ways like these to investigate and facilitate the prevention of cardiovascular toxicity is a rapidly progressing area, and there are a growing number of research articles highlighting hiPSCs in the modeling of anthracyclines, specifically.
Modeling HER2 Monoclonal Antibodies
HER2 is a receptor tyrosine kinase (RTK) that regulates intracellular signaling pathways within various cell types. Trastuzumab and pertuzumab are HER2 monoclonal antibodies that are commonly used to treat breast cancer. RTKs are enzymes that activate proteins via transfer of a phosphate group to a tyrosine residue in a signal transduction cascade. Cancers can overexpress RTKs on their cell surfaces, leading to amplified intracellular signaling, cell proliferation, and tumor initiation [81]. HER2 overexpression is a hallmark of breast cancer and is found in approximately 15 to 20% of all invasive breast cancers [82]. Its proliferation on tumor cells has led to targeted drug therapeutics, including trastuzumab and pertuzumab [83]. Though from the same drug class, these two antibodies (HER2-mAbs) differ in their epitope binding site on the receptor, but their mechanistic action to downregulate intracellular signaling and stimulate a type 1 immune (Th1) response through FcγRIIa and FcγRIIIa activation of cytotoxic cells is similar [84]. In addition, margetuximab, another HER2-mAb with a higher affinity for activating a Th1 response, is also being investigated to treat HER2-positive breast cancer in addition to HER2-positive gastroesophageal adenocarcinoma [85]. Cardiovascular toxicity, especially cardiac dysfunction defined as reduced LVEF, manifests as congestive heart failure (CHF) and is a common side effect of trastuzumab. Underlying mechanisms are still being investigated [86]. The hiPSC models are a promising development in this space that will allow study groups to more clearly identify specific cardiotoxic mechanisms of HER2-mAb therapies and could enable the prediction of susceptibility to cardiotoxicity or offer potential therapies for cardioprotection [87]. Toxicity and adverse effects of margetuximab are still under investigation, though promising clinical trials in patients report little to no adverse effects with the most serious being anemia or infusion-related reactions [88].
Table 1 provides a summary of studies that attempt to identify specific cardiotoxic mechanisms of trastuzumab and predict susceptibility to cardiovascular toxicity. One group evaluated underlying cellular mechanisms of trastuzumab cellular toxicity using hiPSC-CMs [89]. In the study, patient-specific hiPSC-CM models were created for patients on trastuzumab therapy, independent of the development of cardiac dysfunction. The study investigators demonstrated that patient-specific hiPSC-CM models can effectively predict a patient’s risk of trastuzumab-induced cardiovascular toxicity [89]. Another group discovered that clinically relevant doses of trastuzumab impaired contractility, Ca2+ handling, mitochondrial function, and energy metabolism pathways [90•]. These findings expanded the previously proposed mechanisms of cardiovascular toxicity, e.g., activation of mitochondrial apoptotic pathways [91, 92], gene alteration of mitochondrial function, and DNA repair [89, 90•]. Hence, hiPSC-CM models can be used in vitro to recapitulate the cardiotoxic responses of individual patients, predict patient outcomes related to drug safety and efficacy, and further help identify the mechanisms of cardiovascular toxicity [72].
Modeling Tyrosine Kinase Inhibitors
Inhibitors of receptor tyrosine kinases (RTKs), also known as TKIs, have drastically improved cancer survival rates via their improved antitumor efficacy. The most common TKIs include imatinib, erlotinib, sunitinib, sorafenib, crizotinib, gefitinib, and dasatinib. These drugs are a widely used chemotherapeutics that are used to treat various malignancies, including chronic myeloid leukemia, renal cell carcinoma, and gastrointestinal stromal tumors. TKIs selectively prevent phosphorylation of tyrosine kinases without interfering with other serine or threonine kinases. TKIs were shown to competitively inhibit 57 of 317 human kinases tested at the clinically relevant dose of 0.1μM [93]. Selective inhibition of RTKs prevents signaling cascades that lead to over-proliferation, angiogenesis, and growth particularly by inhibiting vascular endothelial growth factor (VEGF) and epidermal growth factor receptor (EGFR) [94,95,96]. These targeted agents have improved antitumor efficacy and fewer toxic side effects than earlier chemotherapeutic agents. Crizotinib is a TKI primarily used to treat non-small cell lung cancer (NSCLC) [34]. It is an inhibitor of the RTK anaplastic lymphoma kinase (ALK) and the proto-oncogene c-Met/hepatocyte growth factor receptor (MET/HGFR) [57]. Erlotinib is generally used for therapy in advanced or metastatic pancreatic or NSCLC and works by inhibiting the activation of EGFR [57]. Nilotinib (a bcr-Abl inhibitor commonly used to treat chronic myeloid leukemia) increases ROS generation and caspase activation and induces arrhythmic beating [34].
Unfortunately, TKIs cause serious adverse cardiovascular toxicity in some patients; this is attributed to the lack of fine-tuned kinase selectivity [93]. Different TKIs show cytotoxicity in hiPSC-CMs, endothelial cells (EC), and fibroblasts (FB), which suggests that TKIs differently affect cardiovascular and non-cardiovascular cell types [93]. Crizotinib induces cholesterol and ROS accumulation and increases caspase activity and in turn increases the S-T interval [34]. Erlotinib generally has more minor effects and does not directly change ROS generation or heart rhythm [34]. When the effects of four different TKIs (erlotinib, lapatinib, sorafenib, and sunitinib) were studied on hiPSC-CMs, each showed slightly different physiology and gene and protein expression. Sorafenib was shown to inhibit mitochondrial ATP production, shifting hiPSCs to rely on glycolysis for energy. This showed that adaptive drug resistance and cardiotoxicity associated with the drug is a consequence of metabolic remodeling instead of inhibition of signal transduction [97].
HiPSCs differentiated into cardiac cells can demonstrate the distinct cardiotoxic effects of different TKIs. The most well-known side effect is hypertension, mainly caused by disruption of the VEGF signaling pathway (VSP) which modulates vasodilation [60]. Up to 47% of patients taking sunitinib and 40% of patients taking sorafenib developed hypertension [60]. TKIs are also documented to cause cardiac left ventricular systolic dysfunction and CHF; 11% of patients taking sunitinib develop CHF or significantly decreased LVEF [96]. Cardiovascular toxicity of TKIs varies widely and depends on the selectivity of the TKIs to certain tissues; sunitinib bound over 15% of kinases included in the human kinase assay [93, 98]. Sunitinib decreased hiPSC-CM viability, increased lipid accumulation, and induced arrhythmic events [34]. Although sunitinib targets multiple RTKs, it was reported that cardiac-specific activation of phosphoinositide 3 kinase (PI3K) could be an effective therapeutic approach to treat sunitinib-induced cardiotoxicity [99]. The VEGF2/platelet-derived growth factor receptor (PDGFR)-inhibiting TKIs can induce cardiovascular toxicities in hiPSCs at clinically relevant concentrations [74], although different TKIs have distinct toxicity profiles; not all TKIs induce cardiovascular toxicity [98]. The pathophysiology of toxicity includes decreased nitric oxide signaling, increased endothelin-1 production, and increased capillary refraction in the endothelium [96]. A long-ranging goal for TKI development should be tissue- or cell-type–specific selectivity and targeting specific kinases in order to reduce cardiovascular toxicity and other global side effects which could be investigated using hiPSCs.
Prevention
At this time, prevention of cardiovascular toxicity from direct and/or indirect damage to the heart from cancer therapies is limited to assessment and optimization of clinical risk factors, taking into account the types of cancers and treatments [2]. Advancements have come with the continuous infusion of a limited dose of liposomal doxorubicin [6]. Anthracycline analogs are being investigated for the treatment of anthracycline-induced toxicity, with hopes of producing less cardiotoxic effects, as well as fine-tuning administration schedules [100]. However, the success of these analogs varies by clinical setting and malignancy, where reductions in cardiovascular toxicity were not always achieved [1]. Many of the cardiotoxic molecular and cellular mechanisms are linked to the highly conserved intracellular kinase: AMPK. It is apparent that this highly conserved intracellular AMPK is at the crucial point of many mechanisms shown to be involved in anthracycline-induced cardiotoxicity, and cardiac AMPK signaling itself has been shown to be impaired by anthracyclines [77]. In a key study, doxorubicin reduced AMPK levels, and AMPK phosphorylation was observed in the heart even before the onset of cardiac dysfunction [101]. Cardioprotection via AMPK activation was achieved in preclinical studies with AMPK-activating agents, such as metformin and sodium–glucose cotransporter 2 inhibitors (SGLT2i) [102]. HER2 activation triggered by exogenous neuregulin-1 could also possibly be used to reduce doxorubicin toxicity [77]. Similar to anthracyclines, trastuzumab toxicity could also be rescued using metabolism-stimulating agents, such as AMPK [90•]. Dexrazoxane is the only FDA-approved agent specifically used to prevent anthracycline-induced cardiovascular toxicity. Though an iron chelator used as a cardioprotective agent when administering anthracyclines, its cancer-related outcomes remain controversial and remain under FDA restrictions for use with patients with metastatic or advanced breast cancer [103].
Typical cardioprotective medications are also employed, including beta blockers, angiotensin-converting enzyme (ACE) inhibitors, angiotensin receptor blockers, statins, and a neprilysin inhibitor. Pharmacologic interventions such as ACE inhibitors (e.g., lisinopril, enalapril, perindopril) have been used to reduce the workload of the heart, along with beta blockers (e.g., carvedilol, metoprolol). Clinical trials are currently being performed to test whether introducing ACE inhibitors concomitantly with anthracycline therapy is more effective than being prescribed after showing evidence of injury due to chemotherapy (NCT03392740, NCT03265574, NCT01968200). Combining anthracyclines with protective compounds could prevent cardiovascular toxicity if the protective agent is able to lower the production of ROS or the workload of the heart [1]. Statins, such as atorvastatin and simvastatin, are also being studied to determine preservation of LVEF and cardiac muscle function along with reducing oxidative stress (NCT01988571, NCT02943590). In addition, studies are also being done observing the effects of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel blockers such as ivabradine on preserving myocardial perfusion and maintaining cardiac contractility despite heart rate reduction in individuals with cancer (NCT03650205).
Lifestyle interventions such as incorporating aerobic or high-intensity interval exercise before, during, or after chemotherapy are being assessed as methods of preventing subsequent cardiac toxicities arising from anthracycline-induced toxicity (NCT02842658, NCT03748550, NCT03131024). These studies are also incorporating remote ischemic conditioning to observe changes in cardiovascular toxicity throughout chemotherapy as well as after the treatment is complete. Cardiac MRI is being used to measure left ventricular cardiac function in studies to determine whether serologic markers (e.g., troponin) can be used to predict cardiovascular toxicities in patients that currently have or previously had cancer. The use of biomarkers may help to identify individuals who may benefit from closer monitoring and those who have lower risk and may not need frequent monitoring; global longitudinal strain (GLS) is also being evaluated [104]. HiPSCs show great potential to be used to understand the physiological effects of cardiovascular toxicity in cancer patients undergoing anthracycline therapy and can be used to assess the long-term effects of pharmacologic therapies administered to cancer patients and allow for better preventive strategies (Table 1). Multiple researchers are dedicating their time to formulating methodologies that provide both cellular integrity and efficiency while also fine-tuning protocols that allow the cells to achieve molecular profiles like that of mature cell types found in vivo [50••, 54, 59].
The preventions and trials discussed above provide a wide range of cardioprotective treatments. However, they are primarily being tested on human subjects and animal models, without incorporating hiPSCs. As discussed in earlier sections, hiPSCs are a robust method of evaluating the risk factors of chemotherapy and have the potential to measure the efficacy of various cardioprotective treatments, such as lifestyle interventions. In the future, these trials should be conducted using hiPSC modeling, which would allow for various prevention combinations to be investigated and eventually personalized risk assessment to be developed. Risk stratification and subsequent prevention measures can then be tailored for patients using their individualized risk factors to determine safety scores, as well as the results from studying the effects of cardioprotective treatments on hiPSCs.
The Future of Modeling Using hiPSCs
Independent of the advancements of oncological treatments themselves, the future of hiPSC modeling will be more dependent on the application of cutting-edge methods and tools that are able to mimic and replicate an individual’s response to treatment in multiple organs and/or cell types. Managing a cell line with one or two cell types can be challenging, but the ambition of many is to create the “perfect” model system—a human-on-a-chip. Although we are not close to that feat, researchers around the globe are experimenting with vascularized, 3D-hiPSC-derived cells, organoids/spheroids, EHT, multi-cell tissue suspension cultures, and even multisensory microfluidic organs-on-a-chip which hold great potential for tissue regeneration and screening drug-induced alterations in cardiac function on a patient-specific level with real-time data collection capabilities [105,106,107,108]. Advanced imaging techniques are also making greater contributions to measurements of chemotherapy-induced cardiovascular toxicity by assessing structural components of hiPSC-derived cardiac cells and microtissues such as ATP depletion and cellular viability, the integrity of the endoplasmic reticulum, and assessments of mitochondrial membrane potential [109••]. Bioinformaticians are also collectively working with clinicians and scientists to analyze, model, and assess risk using these types of data for individuals or populations for better predictive and preventative healthcare [110]. Machine learning, neural networks, and artificial intelligence (AI) are playing larger roles in precision cardio-oncology investigations [111]. With the shear amount of information and data we are now collecting, it is imperative that we have the mathematical and bioinformatic means to properly gather, normalize, analyze, and interpret these data, for which AI is a logical fit.
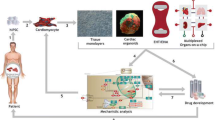
Taken together, the types of omics data and paired phenotypic and environmental data may be able to infer a causal direction and effect for the observed association of a modifiable risk factor and the clinically relevant outcome in question, leading to the discovery of gene–environment interactions based on the variants of interest. hiPSC-based disease modeling is currently a rapidly developing area that has contributions from many disease profiles, especially chemotherapy-induced cardiovascular toxicity. The task of acquiring cells from a patient in a minimally invasive manner, reprogramming the cells into hiPSCs, differentiating them into desired cell type(s) of interest, performing toxicity screens, and extrapolating a patient’s risk of developing cardiotoxicity using defined biological and molecular methodologies (Fig. 1) is where the field of precision cardio-oncology is heading. The future will encompass the identification of biochemical markers and validation studies to confirm current mechanisms of disease and toxicity and identify new mechanisms, screening therapeutic drugs in patient hiPSC-derived cells, and use of cell therapy, grafting, or transplant without the immunological, acute, or chronic adverse effects. It may be plausible that tissue and cell banking will become a routine of every patient where hiPSCs, hiPSC-CMs, or EHT, for example, are readily available when needed by the patient themselves or in massive toxicity screens [112]. Scaling of these types of tissues and cells for population retrospective or prospective studies using forward or reverse genetics and molecular profiling will likely require a global agreement of sorts. It will be imperative to agree on standardized operating procedures, quality assurance, and quality control of many aspects of the reprogramming, culturing, and differentiation methods to ensure maximal reproducibility, purity, and safety, all while doing so in a cost-efficient and time-saving manner. Precise control and regulation of hiPSC-based in vivo therapies will be necessary to prevent tumor formation by overactive cells. A great example will be of hiPSCs with engineered artificial receptors and surrogate antigens (low cost) that can trigger endogenous signal transduction pathways in a dose-dependent manner. STAT3 was activated through signal cascade by an engineered artificial receptor which prevented heart failure and guided hiPSC-CMs into more mature CMs upon doxorubicin administration [113].
Personalized and predictive health assessment using human-induced pluripotent stem cells (hiPSCs). 1, Patient diagnosis of cancer necessitating therapy with substantial potential for cardiovascular toxicity; 2, primary somatic cell types such as peripheral blood mononuclear cells (PBMCs) or fibroblasts taken from the patient to form their own cell line; 3, cultivating and differentiating hiPSCs to cardiomyocytes; 4, studying hiPSC-CM cellular response to chemotherapeutic agents; 5, individualized risk predictions based on cardiotoxicity risk profiles developed using patient’s own hiPSCs; 6, personalized therapy recommended for the patient; 7, shared decision-making (SDM) between the patient and clinician regarding cancer therapy considerations and appropriate cardioprotective measures
Recently, hiPSC-CMs were used to study the mechanisms of cardiovascular toxicity related to coronavirus disease of 2019 (COVID-19) infection and also demonstrate the cytopathic effects, including apoptosis and changes in contractility [114]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection of the cardiovascular system involves the angiotensin-converting enzyme 2 (ACE2), which plays a role in heart function [114]. Knowledge is constantly evolving, but it appears that those with comorbidities, including CVD and cancer, are at increased risk of COVID-19 and having increased risk of mortality [115]. This risk would be even higher in cardio-oncology patients with cardiovascular toxicity due to weakened immune systems and the presence of multiple risk factors [115,116,117]. Continued modeling of hiPSC-CMs may allow for the discovery of pharmaceutical treatment or prevention methods that would especially benefit patients with chemotherapeutic induced cardiovascular toxicity, including in the setting of COVID-19.
Challenges and Limitations of Future Application of hiPSCs
The hiPSC-CM model recapitulates many of the physiological features of drug-induced cardiotoxicity observed in humans, suggesting that it may contribute to physiologically accurate diagnosis of the molecular mechanisms responsible for cardiovascular toxicity. Proof-of-concept studies using hiPSC-CMs to recapitulate the development of cardiotoxicity in cancer patients after doxorubicin treatment and small molecule tyrosine kinase inhibitors have ushered in the promise of iPSC-based disease modeling for toxicity screening. However, previous studies were mostly retrospective on patient samples collected after review of medical charts. By design, retrospective studies must be cautiously interpreted and extrapolated due to limitations of selection bias. There is a need to randomly and prospectively screen and evaluate patient-derived, toxicological profiles using hiPSC technology.
Currently hiPSCs are being used in clinical trials to model disease phenotypes, such as cardiomyopathy, cardiac arrhythmias, and heart failure in the general population (NCT03696628, NCT02413450, NCT03763136). Some of these pathologies have similarities with cardiac damage seen in cardiovascular toxicities from cancer therapies, suggesting potential utility of similar techniques for future cardio-oncology studies. In another clinical trial, 3D-engineered heart tissue derived from hiPSCs for early heart failure detection is being used to analyze individual patient response to known stressors and eventually create a clinically applicable test personalized as avatars for each patient (NCT02417311). While there are many limitations to this trial, the ongoing study elucidates the possibility of a similar system being used in cardio-oncology. To date, there is one clinical trial using hiPSCs as a platform to investigate the cardiotoxic effects of anthracyclines and trastuzumab (NCT03199300). A more comprehensive portfolio of cardio-oncology studies using hiPSCs will be needed to evaluate well-known cardiotoxic chemotherapeutics, investigating the interplay among risk factors, prevention methods, and therapeutic approaches, as well as development of an effective scoring system.
To perform evidence-based studies that could change current practice recommendations for cardio-oncology practice, we need future large-scale pilot studies for the development of personalized risk assessment. These would require the creation of an intervention and randomization of cancer patients and survivors to either standard care versus the use of hiPSCs and evaluation of the necessary clinical endpoints. Future studies are also needed to inform the appropriate design of experiments to assess “responders” vs. “non-responders” of FDA-approved cardiovascular disease therapies, using hiPSCs. Additionally, future studies should address whether other hiPSC-derived lineages (i.e., smooth muscle cells, endothelial cells, fibroblasts), alone or in combination with CMs, can inform cardioprotective strategies related to vascular dysfunction and thrombotic events of chemotherapy-induced cardiotoxicity in humans. Finally, collaboration among multidisciplinary providers of cardiology and medical oncology in partnership with patients could improve care coordination and yield hypothesis-driven clinical studies that impact cardiovascular toxicity in the combined fields of comparative effective research and implementation science. We are confident of overcoming the work’s inherent complexity and level of difficulty to achieve its innovative and highly meritorious strengths in the cardio-oncology field.
For future incorporation in clinical practice, we propose advancement of investigations and overcoming barriers to facilitate using hiPSCs for personalized and predictive health assessment (Fig. 1). The ideal process would begin once the patient first receives their cancer diagnosis. If their prospective therapy regimen requires treatment with anthracyclines, TKIs, or monoclonal antibodies (or other drugs) with known cardiovascular toxicity potential, patients should consider having their individual cell line further analyzed. The analysis would require the patient to provide a sample using either peripheral blood mononuclear cells or fibroblasts, which would be cultivated and differentiated into hiPSC-CMs [48]. The hiPSC-CMs would then be exposed to the patient’s proposed therapies and be observed for cellular responses that are indicative of cardiovascular toxicity. The physician would then bring these results to the patient, along with the recommendation to either continue forward with the initial treatment plan or make the necessary adjustments to prevent cardiovascular toxicity, including using different therapeutic drugs and adding cardioprotective measures. This system would be expected to uncover cardiovascular toxicity predisposition prior to onset by utilizing a relevant human-based, renewable model system, which would ideally allow for more targeted preventative measures and therapeutic modifications.
While this is our hope for the future, we recognize that there are many barriers that first need to be addressed before implementing this system. Practical implementation will be important to consider before using hiPSCs will be ready for clinical practice. First, what proportion of patients will be willing to have their cells obtained and maintained? Second, how much time will this add to the patient experience? Third, for how long will this remain under the auspices of research versus mainstream clinical cardio-oncology? Fourth, is the turnaround time feasible for personalized prediction before therapy starts? Fifth, will there be ambiguity in interpreting results and how will these be adjudicated? Sixth, to what extent will the validity and reliability of the hiPSCs as patient avatars be acceptable? Seventh, how soon would the government, private, and self-insurance entities be ready to reimburse the use of hiPSCs? Eighth, will hiPSCs be widely accessible across public and private healthcare systems in rural, urban, and sub-urban settings with minimal risk of bias and limited access for disadvantaged populations? Providing patients with individualized safety scores based on their on hiPSCs would require ensuring the availability of resources to smaller hospitals and lower-income families.
Indeed, hiPSC-CMs approaches have some additional limitations. hiPSC-CMs are close to the fetal stage of development, and differentiation protocols create a mixture of cells atrial-, ventricular-, and nodal-like phenotypes. In addition, derived CMs are single-cell models and lack three-dimensional tissue interactions of native myocardium. As differentiation protocols advance, including maturation techniques and 3D cultures is likely to overcome some of these problems. Therefore, in the future, there will likely be increased utility of hiPSC-CMs in human disease modeling, high-throughput screening, drug discovery, and prediction of cardiotoxicity.
The applications of hiPSCs can then expand beyond their role in preventing cardiotoxicity to include their use of testing the efficacies of cancer therapies early in the research process, while optimizing cardioprotection. If companies know which drugs are more effective compared to others, and yet still cardioprotective, they can shift their resources, reduce drug withdrawal and save money on costly clinical trials, and reduce patient morbidity and mortality [3]. This would be especially useful with the current state of the world due to SARS-CoV-2. Rapid pharmaceutical discovery and treatment options would be lifesaving, especially for cancer patients who are at increased risk of mortality from viral infections. Further studies utilizing HiPSC modeling should also be considered to help evaluate known cardiovascular risk factors including blood pressure, body mass index, smoking status, diabetes, and physical activity and their contributing roles in the development of cardiovascular toxicity after cancer therapy, to help limit and prevent adverse cardiovascular effects in cancer patients and survivors. Precision cardio-oncology has been in the process of integrating science and medicine in the fields of cardiology and oncology; however, these relationships will intensify in the future and will rely more on systems biology driven by multi-faceted, collaborative (team) approaches to modeling cancer therapy-induced cardiotoxicity.
Conclusion
While innovations in cancer therapeutics have improved survival rates, the associated cardiovascular toxicity is sobering. Cancer and CVD contribute to much of the morbidity and mortality around the world, and it remains important to pursue efforts to manage and prevent cardiovascular toxicity [118]. HiPSC differentiation advancements are allowing researchers to get closer to fully mature cardiomyocytes that can be used to model personalized drug toxicity and disease pathophysiology with increased physiological accuracy. Additional investigation into perfecting multi-cell tissue cultures would aid in identifying the underlying mechanisms of cardiotoxicity and optimize individualized cardiovascular toxicity testing in precision cardio-oncology. Challenges, limitations, and barriers will need to continue to be addressed to create space and pave a path forward for the use of hiPSC in preventive clinical practice in cardio-oncology.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Schwach V, Slaats RH, Passier R. Human pluripotent stem cell-derived cardiomyocytes for assessment of anticancer drug-induced cardiotoxicity. Front Cardiovasc Med. 2020;7(50).
Han X, Zhou Y, Liu W. Precision cardio-oncology: understanding the cardiotoxicity of cancer therapy. NPJ Precis Oncol. 2017;1(1):31.
Stack JP, Moslehi J, Sayed N, Wu JC. Cancer therapy-induced cardiomyopathy: can human induced pluripotent stem cell modelling help prevent it? Eur Heart J. 2018;40(22):1764–70.
Brown SA, Sandhu N, Herrmann J. Systems biology approaches to adverse drug effects: the example of cardio-oncology. Nat Rev Clin Oncol. 2015;12(12):718–31.
Brown SA, Nhola L, Herrmann J. Cardiovascular toxicities of small molecule tyrosine kinase inhibitors: an opportunity for systems-based approaches. Clin Pharmacol Ther. 2017;101(1):65–80.
Brown SA. Preventive cardio-oncology: the time has come. Front Cardiovasc Med. 2019;6:187.
Campia U, Moslehi JJ, Amiri-Kordestani L, Barac A, Beckman JA, Chism DD, et al. Cardio-oncology: vascular and metabolic perspectives: a scientific statement from the American Heart Association. Circulation. 2019;139(13):e579–602.
Blaes AH, Thavendiranathan P, Moslehi J. Cardiac toxicities in the era of precision medicine: underlying risk factors, targeted therapies, and cardiac biomarkers. Am Soc Clin Oncol Educ Book. 2018;38:764–74.
Mehta LS, Watson KE, Barac A, Beckie TM, Bittner V, Cruz-Flores S, et al. Cardiovascular disease and breast cancer: where these entities intersect: a scientific statement from the American Heart Association. Circulation. 2018;137(8):e30–66.
Handy CE, Quispe R, Pinto X, Blaha MJ, Blumenthal RS, Michos ED, et al. Synergistic opportunities in the interplay between cancer screening and cardiovascular disease risk assessment: together we are stronger. Circulation. 2018;138(7):727–34.
Moslehi JJ. Cardiovascular toxic effects of targeted cancer therapies. N Engl J Med. 2016;375(15):1457–67.
Pavo N, Arfsten H, Cho A, Goliasch G, Bartko PE, Wurm R, et al. The circulating form of neprilysin is not a general biomarker for overall survival in treatment-naïve cancer patients. Sci Rep. 2019;9(1):2554.
Gilliland TM, Villafane-Ferriol N, Shah KP, Shah RM, Tran Cao HS, Massarweh NN, et al. Nutritional and metabolic derangements in pancreatic cancer and pancreatic resection. Nutrients. 2017;9(3).
Tonorezos ES, Jones LW. Energy balance and metabolism after cancer treatment. Semin Oncol. 2013;40(6):745–56.
van Waas M, Neggers SJ, Raat H, van Rij CM, Pieters R, van den Heuvel-Eibrink MM. Abdominal radiotherapy: a major determinant of metabolic syndrome in nephroblastoma and neuroblastoma survivors. PLoS One. 2012;7(12):e52237.
Gibson TM, Ehrhardt MJ, Ness KK. Obesity and metabolic syndrome among adult survivors of childhood leukemia. Curr Treat Options in Oncol. 2016;17(4):17.
Gurney JG, Kadan-Lottick NS, Packer RJ, Neglia JP, Sklar CA, Punyko JA, et al. Endocrine and cardiovascular late effects among adult survivors of childhood brain tumors: childhood cancer survivor Study. Cancer. 2003;97(3):663–73.
DeSantis CE, Siegel RL, Sauer AG, Miller KD, Fedewa SA, Alcaraz KI, et al. Cancer statistics for African Americans, 2016: progress and opportunities in reducing racial disparities. CA Cancer J Clin. 2016;66(4):290–308.
Hasan S, Dinh K, Lombardo F, Kark J. Doxorubicin cardiotoxicity in African Americans. J Natl Med Assoc. 2004;96(2):196–9.
Lotrionte M, Biondi-Zoccai G, Abbate A, Lanzetta G, D'Ascenzo F, Malavasi V, et al. Review and meta-analysis of incidence and clinical predictors of anthracycline cardiotoxicity. Am J Cardiol. 2013;112(12):1980–4.
Finkelman BS, Putt M, Wang T, Wang L, Narayan H, Domchek S, et al. Arginine-nitric oxide metabolites and cardiac dysfunction in patients with breast cancer. J Am Coll Cardiol. 2017;70(2):152–62.
Litvak A, Batukbhai B, Russell SD, Tsai HL, Rosner GL, Jeter SC, et al. Racial disparities in the rate of cardiotoxicity of HER2-targeted therapies among women with early breast cancer. Cancer. 2018;124(9):1904–11.
Baron KB, Brown JR, Heiss BL, Marshall J, Tait N, Tkaczuk KH, et al. Trastuzumab-induced cardiomyopathy: incidence and associated risk factors in an inner-city population. J Card Fail. 2014;20(8):555–9.
Özdemir BC, Dotto GP. Racial differences in cancer susceptibility and survival: more than the color of the skin? Trends Cancer. 2017;3(3):181–97.
Prasad P, Branch M, Asemota D, Elsayed R, Addison D, Brown S-A. Cardio-oncology preventive care: racial and ethnic disparities. Curr Cardiovasc Risk Rep. 2020;14(10):18.
• Dent SF, Kikuchi R, Kondapalli L, Ismail-Khan R, Brezden-Masley C, Barac A, et al. Optimizing cardiovascular health in patients with cancer: a practical review of risk assessment, monitoring, and prevention of cancer treatment-related cardiovascular toxicity. Am Soc Clin Oncol Educ Book. 2020;40:1–15 Discusses the risks of therapies on cardiovascular toxicity. Furthermore, describes how cardiac risk factors are associated with cancer, as well as how cardiac risk factors and cancers are associated with worsening cardiac disease. Also discusses cardiac monitoring strategies.
Octavia Y, Tocchetti CG, Gabrielson KL, Janssens S, Crijns HJ, Moens AL. Doxorubicin-induced cardiomyopathy: from molecular mechanisms to therapeutic strategies. J Mol Cell Cardiol. 2012;52(6):1213–25.
Liang P, Lan F, Lee AS, Gong T, Sanchez-Freire V, Wang Y, et al. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation. 2013;127(16):1677–91.
Sayed N, Ameen M, Wu JC. Personalized medicine in cardio-oncology: the role of induced pluripotent stem cell. Cardiovasc Res. 2019;115(5):949–59.
• Abdel-Qadir H, Thavendiranathan P, Austin PC, Lee DS, Amir E, Tu JV, et al. Development and validation of a multivariable prediction model for major adverse cardiovascular events after early stage breast cancer: a population-based cohort study. Eur Heart J. 2019;40(48):3913–20 Authors created a scoring system to predict major cardiovascular events after early-stage breast cancer. This scoring system used age, past medical history of heart failure, atrial fibrillation, peripheral vascular disease, hypertension, ischemic heart disease, diabetes, chronic kidney disease, COPD, and cerebrovascular disease in order to risk stratify patients for risk at 5 years and at 10 years.
Herrmann J, Lerman A, Sandhu NP, Villarraga HR, Mulvagh SL, Kohli M. Evaluation and management of patients with heart disease and cancer: cardio-oncology. Mayo Clin Proc. 2014;89(9):1287–306.
Karakikes I, Ameen M, Termglinchan V, Wu JC. Human induced pluripotent stem cell-derived cardiomyocytes: insights into molecular, cellular, and functional phenotypes. Circ Res. 2015;117(1):80–8.
Genova E, Cavion F, Lucafò M, Leo L, Pelin M, Stocco G, et al. Induced pluripotent stem cells for therapy personalization in pediatric patients: focus on drug-induced adverse events. World J Stem Cells. 2019;11(12):1020–44.
Doherty KR, Wappel RL, Talbert DR, Trusk PB, Moran DM, Kramer JW, et al. Multi-parameter in vitro toxicity testing of crizotinib, sunitinib, erlotinib, and nilotinib in human cardiomyocytes. Toxicol Appl Pharmacol. 2013;272(1):245–55.
Talbert DR, Doherty KR, Trusk PB, Moran DM, Shell SA, Bacus S. A multi-parameter in vitro screen in human stem cell-derived cardiomyocytes identifies ponatinib-induced structural and functional cardiac toxicity. Toxicol Sci. 2015;143(1):147–55.
Liang M. Epigenetic mechanisms and hypertension. Hypertension. 2018;72(6):1244–54.
Mishra MK, Liang EY, Geurts AM, Auer PWL, Liu P, Rao S, et al. Comparative and functional genomic resource for mechanistic studies of human blood pressure-associated single nucleotide polymorphisms. Hypertension. 2020;75(3):859–68.
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72.
Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–20.
Wang AYL, Loh CYY. Episomal induced pluripotent stem cells: functional and potential therapeutic applications. Cell Transplant. 2019;28(1_suppl):112S–31S.
Churko JM, Lee J, Ameen M, Gu M, Venkatasubramanian M, Diecke S, et al. Transcriptomic and epigenomic differences in human induced pluripotent stem cells generated from six reprogramming methods. Nat Biomed Eng. 2017;1(10):826–37.
Weber M, Schübeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr Opin Cell Biol. 2007;19(3):273–80.
Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196(2):261–82.
•• Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29(12):1117–9 An older, but relevant, publication that discusses the importance of donor cell type choice and the impact it may have on reprogramming process and subsequent differentiation into desired cell types. Profiling differentiated cell types using molecular methods is shaping how we view overall cell maturity and proper function.
Moretti A, Laugwitz KL, Dorn T, Sinnecker D, Mummery C. Pluripotent stem cell models of human heart disease. Cold Spring Harb Perspect Med. 2013;3(11).
Doyle MJ, Lohr JL, Chapman CS, Koyano-Nakagawa N, Garry MG, Garry DJ. Human induced pluripotent stem cell-derived cardiomyocytes as a model for heart development and congenital heart disease. Stem Cell Rev Rep. 2015;11(5):710–27.
Yang X, Rodriguez ML, Leonard A, Sun L, Fischer KA, Wang Y, et al. Fatty acids enhance the maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cell Rep. 2019;13(4):657–68.
Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11(8):855–60.
Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109(27):E1848–57.
•• Feyen DAM, McKeithan WL, Bruyneel AAN, Spiering S, Hormann L, Ulmer B, et al. Metabolic maturation media improve physiological function of human iPSC-derived cardiomyocytes. Cell Rep. 2020;32(3):107925 This is a very thorough study into how alterations in the media for hiPSC-CMs can provide a better model system by elevating the physiological maturity level. Multifaceted approaches are aiding in bettering the hiPSC models for drug toxicity screenings and drug discovery, leading to more precision in diagnoses and care. Small mechanistic details in a protocol may have great impacts on the model system.
Horikoshi Y, Yan Y, Terashvili M, Wells C, Horikoshi H, Fujita S, et al. Fatty acid-treated induced pluripotent stem cell-derived human cardiomyocytes exhibit adult cardiomyocyte-like energy metabolism phenotypes. Cells. 2019;8(9).
Hazeltine LB, Simmons CS, Salick MR, Lian X, Badur MG, Han W, et al. Effects of substrate mechanics on contractility of cardiomyocytes generated from human pluripotent stem cells. Int J Cell Biol. 2012;2012:508294.
Lian X, Bao X, Zilberter M, Westman M, Fisahn A, Hsiao C, et al. Chemically defined, albumin-free human cardiomyocyte generation. Nat Methods. 122015:595–6.
Buikema JW, Lee S, Goodyer WR, Maas RG, Chirikian O, Li G, et al. Wnt activation and reduced cell-cell contact synergistically induce massive expansion of functional human iPSC-derived cardiomyocytes. Cell Stem Cell. 2020;27(1):50–63 e5.
Floy ME, Mateyka TD, Foreman KL, Palecek SP. Human pluripotent stem cell-derived cardiac stromal cells and their applications in regenerative medicine. Stem Cell Res. 2020;45:101831.
Bao X, Lian X, Qian T, Bhute VJ, Han T, Palecek SP. Directed differentiation and long-term maintenance of epicardial cells derived from human pluripotent stem cells under fully defined conditions. Nat Protoc. 2017;12(9):1890–900.
Zhang H, Shen M, Wu JC. Generation of quiescent cardiac fibroblasts derived from human induced pluripotent stem Cells. Methods Mol Biol. 2020.
Chen Z, Xian W, Bellin M, Dorn T, Tian Q, Goedel A, et al. Subtype-specific promoter-driven action potential imaging for precise disease modelling and drug testing in hiPSC-derived cardiomyocytes. Eur Heart J. 2017;38(4):292–301.
Dunn KK, Reichardt IM, Simmons AD, Jin G, Floy ME, Hoon KM, et al. Coculture of endothelial cells with human pluripotent stem cell-derived cardiac progenitors reveals a differentiation stage-specific enhancement of cardiomyocyte maturation. Biotechnol J. 2019;14(8):e1800725.
Wang K, Lin R-Z, Hong X, Ng AH, Lee CN, Neumeyer J, et al. Robust differentiation of human pluripotent stem cells into endothelial cells via temporal modulation of ETV2 with modified mRNA. Sci Adv. 2020;6(30):eaba7606.
Gewirtz DA. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem Pharmacol. 1999;57(7):727–41.
Bailly C. Contemporary challenges in the design of topoisomerase II inhibitors for cancer chemotherapy. Chem Rev. 2012;112(7):3611–40.
Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9(5):338–50.
Gratia S, Kay L, Potenza L, Seffouh A, Novel-Chaté V, Schnebelen C, et al. Inhibition of AMPK signalling by doxorubicin: at the crossroads of the cardiac responses to energetic, oxidative, and genotoxic stress. Cardiovasc Res. 2012;95(3):290–9.
Ferreira AL, Matsubara LS, Matsubara BB. Anthracycline-induced cardiotoxicity. Cardiovasc Hematol Agents Med Chem. 2008;6(4):278–81.
McGowan JV, Chung R, Maulik A, Piotrowska I, Walker JM, Yellon DM. Anthracycline chemotherapy and cardiotoxicity. Cardiovasc Drugs Ther. 2017;31(1):63–75.
Finck BN, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) regulatory cascade in cardiac physiology and disease. Circulation. 2007;115(19):2540–8.
Lyu YL, Kerrigan JE, Lin CP, Azarova AM, Tsai YC, Ban Y, et al. Topoisomerase IIbeta mediated DNA double-strand breaks: implications in doxorubicin cardiotoxicity and prevention by dexrazoxane. Cancer Res. 2007;67(18):8839–46.
Zhang S, Liu X, Bawa-Khalfe T, Lu L-S, Lyu YL, Liu LF, et al. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat Med. 2012;18(11):1639–42.
Magdy T, Schuldt AJT, Wu JC, Bernstein D, Burridge PW. Human induced pluripotent stem cell (hiPSC)-derived cells to assess drug cardiotoxicity: opportunities and problems. Annu Rev Pharmacol Toxicol. 2018;58:83–103.
Nair P, Prado M, Perea-Gil I, Karakikes I. Concise review: precision matchmaking: induced pluripotent stem cells meet cardio-oncology. Stem Cells Transl Med. 2019;8(8):758–67.
Burridge PW, Li YF, Matsa E, Wu H, Ong SG, Sharma A, et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med. 2016;22(5):547–56.
Louisse J, Wüst RCI, Pistollato F, Palosaari T, Barilari M, Macko P, et al. Assessment of acute and chronic toxicity of doxorubicin in human induced pluripotent stem cell-derived cardiomyocytes. Toxicol in Vitro. 2017;42:182–90.
Sharma A, Burridge PW, McKeithan WL, Serrano R, Shukla P, Sayed N, et al. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci Transl Med. 2017;9(377).
Cohen JD, Babiarz JE, Abrams RM, Guo L, Kameoka S, Chiao E, et al. Use of human stem cell derived cardiomyocytes to examine sunitinib mediated cardiotoxicity and electrophysiological alterations. Toxicol Appl Pharmacol. 2011;257(1):74–83.
Kurokawa YK, Shang MR, Yin RT, George SC. Modeling trastuzumab-related cardiotoxicity in vitro using human stem cell-derived cardiomyocytes. Toxicol Lett. 2018;285:74–80.
Eldridge S, Guo L, Mussio J, Furniss M, Hamre J, Davis M. Examining the protective role of ErbB2 modulation in human-induced pluripotent stem cell-derived cardiomyocytes. Toxicol Sci. 2014;141(2):547–59.
Gupta SK, Garg A, Bar C, Chatterjee S, Foinquinos A, Milting H, et al. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ Res. 2018;122(2):246–54.
Weng K-C, Kurokawa YK, Hajek BS, Paladin JA, Shirure VS, George SC. Human induced pluripotent stem-cardiac-endothelial-tumor-on-a-chip to assess anticancer efficacy and cardiotoxicity. Tissue Eng C Methods. 2020;26(1):44–55.
Christidi E, Huang H, Shafaattalab S, Maillet A, Lin E, Huang K, et al. Variation in RARG increases susceptibility to doxorubicin-induced cardiotoxicity in patient specific induced pluripotent stem cell-derived cardiomyocytes. Sci Rep. 2020;10(1):10363.
Hsu JL, Hung M-C. The role of HER2, EGFR, and other receptor tyrosine kinases in breast cancer. Cancer Metastasis Rev. 2016;35(4):575–88.
Ng CK, Martelotto LG, Gauthier A, Wen H-C, Piscuoglio S, Lim RS, et al. Intra-tumor genetic heterogeneity and alternative driver genetic alterations in breast cancers with heterogeneous HER2 gene amplification. Genome Biol. 2015;16(1):107.
Montemurro F, Scaltriti M. Biomarkers of drugs targeting HER-family signalling in cancer. J Pathol. 2014;232(2):219–29.
Costa RL, Czerniecki BJ. Clinical development of immunotherapies for HER2+ breast cancer: a review of HER2-directed monoclonal antibodies and beyond. NPJ Breast Cancer. 2020;6(1):1–11.
Rugo HS, Im S-A, Wright GLS, Escriva-de-Romani S, DeLaurentiis M, Cortes J, et al. SOPHIA primary analysis: a phase 3 (P3) study of margetuximab (M)+ chemotherapy (C) versus trastuzumab (T)+ C in patients (pts) with HER2+ metastatic (met) breast cancer (MBC) after prior anti-HER2 therapies (Tx). Proc Am Soc Clin Oncol. 2019.
Nemeth BT, Varga ZV, Wu WJ, Pacher P. Trastuzumab cardiotoxicity: from clinical trials to experimental studies. Br J Pharmacol. 2017;174(21):3727–48.
Hahn VS, Lenihan DJ, Ky B. Cancer therapy-induced cardiotoxicity: basic mechanisms and potential cardioprotective therapies. J Am Heart Assoc. 2014;3(2):e000665.
Catenacci DV, Kang Y-K, Park H, Uronis HE, Lee K-W, Ng MC, et al. Margetuximab plus pembrolizumab in patients with previously treated, HER2-positive gastro-oesophageal adenocarcinoma (CP-MGAH22–05): a single-arm, phase 1b–2 trial. Lancet Oncol. 2020;21(8):1066–76.
Mohan N, Jiang J, Dokmanovic M, Wu WJ. Trastuzumab-mediated cardiotoxicity: current understanding, challenges, and frontiers. Antibody Ther. 2018;1(1):13–7.
• Kitani T, Ong S-G, Lam CK, Rhee J-W, Zhang JZ, Oikonomopoulos A, et al. Human-induced pluripotent stem cell model of trastuzumab-induced cardiac dysfunction in patients with breast cancer. Circulation. 2019;139(21):2451–65 Researchers used hiPSC-CM models to further investigate the cellular mechanism of action of the cardiotoxic effects of trastuzumab. Patient models of hiPSC-CM were successfully shown to recapitulate the same effects in vitro.
Grazette LP, Boecker W, Matsui T, Semigran M, Force TL, Hajjar RJ, et al. Inhibition of ErbB2 causes mitochondrial dysfunction in cardiomyocytes: implications for herceptin-induced cardiomyopathy. J Am Coll Cardiol. 2004;44(11):2231–8.
Singh KK, Shukla PC, Quan A, Lovren F, Pan Y, Wolfstadt JI, et al. Herceptin, a recombinant humanized anti-ERBB2 monoclonal antibody, induces cardiomyocyte death. Biochem Biophys Res Commun. 2011;411(2):421–6.
Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26(1):127–32.
Di Lorenzo G, Autorino R, Sternberg CN. Metastatic renal cell carcinoma: recent advances in the targeted therapy era. Eur Urol. 2009;56(6):959–71.
Schmidinger M, Zielinski CC, Vogl UM, Bojic A, Bojic M, Schukro C, et al. Cardiac toxicity of sunitinib and sorafenib in patients with metastatic renal cell carcinoma. J Clin Oncol. 2008;26(32):5204–12.
Chu TF, Rupnick MA, Kerkela R, Dallabrida SM, Zurakowski D, Nguyen L, et al. Cardiotoxicity associated with tyrosine kinase inhibitor sunitinib. Lancet. 2007;370(9604):2011–9.
Wang H, Sheehan RP, Palmer AC, Everley RA, Boswell SA, Ron-Harel N, et al. Adaptation of human iPSC-derived cardiomyocytes to tyrosine kinase inhibitors reduces acute cardiotoxicity via metabolic reprogramming. Cell Syst. 2019;8(5):412–26.e7.
Force T, Krause DS, Van Etten RA. Molecular mechanisms of cardiotoxicity of tyrosine kinase inhibition. Nat Rev Cancer. 2007;7(5):332–44.
Li C, Zou R, Zhang H, Wang Y, Qiu B, Qiu S, et al. Upregulation of phosphoinositide 3-kinase prevents sunitinib-induced cardiotoxicity in vitro and in vivo. Arch Toxicol. 2019;93(6):1697–712.
Payne DL, Nohria A. Prevention of chemotherapy induced cardiomyopathy. Curr Heart Fail Rep. 2017;14(5):398–403.
Tokarska-Schlattner M, Zaugg M, Da Silva R, Lucchinetti E, Schaub MC, Wallimann T, et al. Acute toxicity of doxorubicin on isolated perfused heart: response of kinases regulating energy supply. Am J Phys Heart Circ Phys. 2005;289(1):H37–47.
Oh C-M, Cho S, Jang J-Y, Kim H, Chun S, Choi M, et al. Cardioprotective potential of an SGLT2 inhibitor against doxorubicin-induced heart failure. Korean Circ J. 2019;49(12):1183–95.
Barbar T, Mahmood SS, Liu JE. Cardiomyopathy prevention in cancer patients. Cardiol Clin. 2019;37(4):441–7.
Chen CL. Cardiovascular prevention in the cancer survivor. Curr Atheroscler Rep. 2015;17(3):484.
Oleaga C, Bernabini C, Smith AS, Srinivasan B, Jackson M, McLamb W, et al. Multi-organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci Rep. 2016;6:20030.
Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Mousavi Shaegh SA, et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci U S A. 2017;114(12):E2293–E302.
Le MNT, Hasegawa K. Expansion culture of human pluripotent stem cells and production of cardiomyocytes. Bioengineering (Basel). 2019;6(2).
Amano Y, Nishiguchi A, Matsusaki M, Iseoka H, Miyagawa S, Sawa Y, et al. Development of vascularized iPSC derived 3D-cardiomyocyte tissues by filtration layer-by-layer technique and their application for pharmaceutical assays. Acta Biomater. 2016;33:110–21.
•• Cao L, der Meer ADV, Verbeek FJ, Passier R. Automated image analysis system for studying cardiotoxicity in human pluripotent stem cell-Derived cardiomyocytes. BMC Bioinform. 2020;21(1):187 Researchers discuss an automated image analysis system that can detect damaged hiPSCs within a heterogenous cardiac cell culture. Being able to quantify the visualized cardiotoxic phenotypes at a single-cell segmentation level using nuclear signals is astounding and a great contribution to the field.
Sachinidis A. Cardiotoxicity and heart failure: lessons from human-induced pluripotent stem cell-derived cardiomyocytes and anticancer drugs. Cells. 2020;9(4).
Kim H, Nam H. hERG-Att: Self-attention-based deep neural network for predicting hERG blockers. Comput Biol Chem. 2020;87:107286.
Huang C-Y, Liu C-L, Ting C-Y, Chiu Y-T, Cheng Y-C, Nicholson MW, et al. Human iPSC banking: barriers and opportunities. J Biomed Sci. 2019;26(1).
Tsukamoto T, Sogo T, Ueyama T, Nakao S, Harada Y, Ihara D, et al. Chimeric G-CSF receptor-mediated STAT3 activation contributes to efficient induction of cardiomyocytes from mouse induced pluripotent stem cells. Biotechnol J. 2020;15(2):1900052.
Sharma A, Garcia G, Wang Y, Plummer JT, Morizono K, Arumugaswami V, et al. Human iPSC-derived cardiomyocytes are susceptible to SARS-CoV-2 infection. Cell Rep Med. 2020;1(4):100052.
Ganatra S, Hammond SP, Nohria A. The novel coronavirus disease (COVID-19) threat for patients with cardiovascular disease and cancer. JACC Cardio Oncol. 2020;2(2):350–5.
Brown SA, Zaharova S, Mason P, Thompson J, Thapa B, Ishizawar D, et al. Pandemic perspective: commonalities between COVID-19 and cardio-oncology. Front Cardiovasc Med. 2020;7:568720.
Asokan I, Rabadia SV, Yang EH. The COVID-19 Pandemic and its impact on the cardio-oncology population. Curr Oncol Rep. 2020;22(6):60.
Mladěnka P, Applová L, Patočka J, Costa VM, Remiao F, Pourová J, et al. Comprehensive review of cardiovascular toxicity of drugs and related agents. Med Res Rev. 2018;38(4):1332–403.
Archer CR, Sargeant R, Basak J, Pilling J, Barnes JR, Pointon A. Characterization and validation of a human 3D cardiac microtissue for the assessment of changes in cardiac pathology. Sci Rep. 2018;8(1).
Pointon A, Pilling J, Dorval T, Wang Y, Archer C, Pollard C. From the cover: high-throughput imaging of cardiac microtissues for the assessment of cardiac contraction during drug discovery. Toxicol Sci. 2017;155(2):444–57.
Parrotta EI, Lucchino V, Scaramuzzino L, Scalise S, Cuda G. Modeling cardiac disease mechanisms using induced pluripotent stem cell-derived cardiomyocytes: progress, promises and challenges. Int J Mol Sci. 2020;21(12):4354.
Pfeffer TJ, Pietzsch S, Hilfiker-Kleiner D. Common genetic predisposition for heart failure and cancer. Herz. 2020.
Li X, Liu J, Lu Q, Ren D, Sun X, Rousselle T, et al. AMPK: a therapeutic target of heart failure—not only metabolism regulation. Biosci Rep. 2019;39(1):BSR20181767.
Acknowledgements
We are grateful to Dr. Ulrich Broeckel and Dr. Ivor Benjamin at the Medical College of Wisconsin for their helpful comments on this manuscript.
Funding
T.P. received funding through the Jeschke Scholarship Fund.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Cardio-oncology
Rights and permissions
About this article
Cite this article
Perry, T.R., Roberts, M.L., Sunkara, B. et al. Modeling Precision Cardio-Oncology: Using Human-Induced Pluripotent Stem Cells for Risk Stratification and Prevention. Curr Oncol Rep 23, 77 (2021). https://doi.org/10.1007/s11912-021-01066-2
Accepted:
Published:
DOI: https://doi.org/10.1007/s11912-021-01066-2