Abstract
Betula luminifera is a commercial tree species that is emerging as a new model system for tree genomics research. A draft genomic sequence is expected to be publicly available in the near future, which means that an explosion of gene expression studies awaits. Thus, the work of selecting appropriate reference genes for qPCR normalization in different tissues or under various experimental conditions is extremely valuable. In this study, ten candidate genes were analyzed in B. luminifera subjected to different abiotic stresses and at various flowering stages. The expression stability of these genes was evaluated using three distinct algorithms implemented using geNorm, NormFinder and BestKeeper. The best-ranked reference genes varied across different sample sets, though RPL39, MDH and EF1a were determined as the most stable by the three programs among all tested samples. RPL39 and EF1a should be appropriate for normalization in N-starved roots, while the combination of RPL39 and MDH should be appropriate for N-starved stems and EF1a and MDH should be appropriate in N-starved leaves. In PEG-treated (osmotic) roots, MDH was the most suitable, whereas EF1a was suitable for PEG-treated stems and leaves. TUA was also stably expressed levels in PEG-treated plants. The combination of RPL39 and TUB should be appropriate for heat-stressed leaves and flowering stage. For reference gene validation, the expression levels of SOD and NFYA-3 were investigated. This work will be beneficial to future studies on gene expression under different abiotic stress conditions and flowering status in B. luminifera.
Similar content being viewed by others
Introduction
Betula luminifera is a deciduous tree, a member of the Betulaceae family, and is widely distributed in the 14 provinces of China. Its good wood properties, beautiful texture and fast-growing character make it widely used in solid wood furniture and flooring. Furthermore, B. luminifera is highly adaptable to barren soil, thus it was proposed as a priority timber species for afforestation (Huang et al. 2014). In addition to its commercial traits, B. luminifera has great potential for use as a species for forest genetic studies, exemplified by its small genome size (approximately 450 Mb) and a comparatively short juvenile period. Most lines of B. luminifera begin flowering at 1.5 years old, and it has the potential to be a model for woody plants that usually have long juvenile periods. It may also shorten the genetic improvement period for tree breeding. Most tree plantations are located on barren lands where the available nitrate is extremely low, limiting plant growth (Bigelow and Canham 2007). Moreover, extreme weather conditions, such as high temperature and drought, occur frequently in these areas and may affect plant development (Guenin et al. 2009). For plants, the subtle evolutionary changes that accompanied the process of plant domestication are exemplified by a series of genes in response to these abiotic stresses (Mittler and Blumwald 2010).
Gene expression analysis is a fundamental approach for functional genomics studies, which can be used to estimate complicated gene regulatory networks (Obrero et al. 2011). Currently, reverse transcription quantitative real-time PCR (RT-qPCR), extremely sensitive, efficient and reproducible method, is frequently used to quantify gene transcript levels and to analyze gene abundance among different plant tissues, developmental stages and abiotic stresses (Chen et al. 2011; Hong et al. 2010; Wan et al. 2010). However, a number of variables are involved in the accuracy of RT-qPCR. Thus, it is indispensable to adopt a normalization step to avoid bias. The most useful approach for normalization is to apply one or several reference genes, whose expressions are presumed to be stable in both control and experimental conditions (Guenin et al. 2009). Extensive transcriptomics mining and experimental data have shown that there is no such universal reference gene that can be used in all plant tissues under all experimental conditions (Chen et al. 2011). Statistical algorithms have been developed recently but, surprisingly, this robust approach is under-utilized in plants. Instead, putative housekeeping genes tend to be used as references without any proper validation, which may lead to misinterpretation of some results (Guenin et al. 2009).
Studies involving the selection of reference genes have increased, but mainly focus on model and important crop species, such as Arabidopsis (Hong et al. 2010), rice (Bustin et al. 2009; Udvardi et al. 2008), cucumber (Wan et al. 2010), soybean (Jian et al. 2008), wheat (Paolacci et al. 2009), and poplar (Brunner et al. 2004). There has been no systematic analysis for the selection of reference genes for RT-qPCR in B. luminifera, which may adversely impact molecular research of this species. In this study, ten candidate reference genes were selected to normalize the results of gene expression studies under nitrate starvation, osmotic stress and high temperature conditions and at differing flowering stages using three algorithms: geNorm, NormFinder and Bestkeeper (Udvardi et al. 2008).
Materials and methods
Plant materials and treatments
The G49-3 line of B. luminifera was used. The seedlings were maintained on half strength Murashige and Skoog (1/2MS) medium with 30 g L−1 sucrose and solidified with 6 g L−1 agar, pH adjusted to 5.9, with 16 h light/8 h dark.
For experimental treatments, the top three internodes of seedlings were placed on bridges made of filter-paper in glass tubes, which were filled with 40 mL of half solid (3 g L−1 agar) 1/2MS medium. After 1 month of growth, only uniformly developed plants were used in subsequent experiments. For the nitrate starvation treatment, the original medium was suctioned out and displaced by non-nitrate medium, and K+ concentration was adjusted by the addition of KCl for 1, 2 or 10 days. For osmotic treatment, plants were placed in PEG6000 (15 %) for 1, 2 or 5 days. For heat stress treatment, seedlings were grown at 45 °C for 0.5, 1, 2 or 4 h. For the analysis of flowering stages, both male and female inflorescences were sampled every 3 days.
Roots, stems and leaves were collected from plants subjected to nitrate starvation and osmotic stress. For the high temperature treatment, only leaves were collected. Three biological replicates were sampled for all treatments, and these materials were immediately frozen in liquid nitrogen and stored at −80 °C for subsequent use.
RNA isolation, purification and cDNA synthesis
Total RNA was extracted from collected samples using Trizol reagent, according to the manufacturer’s instructions (Invitrogen). RNA samples were treated with RNase-free DNase (Promega) to remove remaining DNA and the integrity was examined using 1 % agarose gel electrophoresis. RNA concentration and purity was determined using a NanoDrop ND-2000 spectrophotometer (Thermo-Scientific, Wilmington, DE, USA). RNA samples with an absorbance ratio at OD260/280 between 1.8 and 2.2 and OD 260/230 of ~2.0 were used for further analysis. Complementary DNA (cDNA) was synthesized using the Primer Script RT reagent kit (TaKaRa Biotechnology, Dalian, China) following the manufacturer’s instructions in a final volume of 20 μL using 1 μg of total RNA. The final cDNA products were diluted fivefold prior to use in qPCR.
Reference gene selection, primer design and amplification efficiency
To identify suitable reference genes for B. luminifera, ten candidates that are frequently used or newly identified in other plant species were selected (Supplementary Table 1). These candidates included such traditional reference genes as large ribosomal subunit 39 (RPL39), polyubiquitin 10 (UBQ10), ribosomal protein S24 (S24), β-tubulin (TUB), malate dehydrogenase (MDH), elongation factor 1-a (EF1-a), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a-tubulin (TUA), actin7 (ACT), and a novel identified stable reference gene of Arabidopsis, the TIP-41 like protein (TIP41).
To obtain B. luminifera orthologous sequences of the candidates, BLASTN searches were conducted using Arabidopsis genes as queries on B. luminifera unigenes in the transcriptomic data. PCR primers were designed using Oligo 7.0 software (Molecular Biological Insights, Colorado Springs, CO, USA) with melting temperatures of 60 ± 1 °C and amplicon lengths of 100–200 bp (Table 1, Supplementary Table 2). Specific products and amplicon lengths of all primer pairs were verified using 2.0 % agarose gel electrophoresis (Supplementary Fig. 1). The amplification efficiency of each primer pair was calculated using a standard curve generated from a serial dilution of cDNA (1, 10, 102, 103, 104 dilutions). Mean quantification cycle (C q) values of each tenfold dilution were plotted against the logarithm of the cDNA dilution factor. The amplification efficiency (E) for each gene was calculated using the slope of a linear regression model (Pfaffl 2001) using the C q values and the following equation: E = [10(1/−slope) − 1] × 100 %.
qPCR and statistical analyses
The transcript abundance for each of the ten genes was analyzed by qPCR (CFX96, BioRad, Hercules, USA) using SYBR Premix EX Taq (TaKaRa Biotechnology, Dalian, China) in a volume of 10 μL, following the manufacturer’s instructions. Thermal conditions were 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 20 s. The dissociation curve was obtained by heating the amplicon from 60 to 95 °C, and the melting curve had a single peak (Supplementary Fig. 2). All qPCR reactions were performed in technical triplicate. In this work, we followed the minimum information for publication of qPCR experiments (MIQE) (Bustin et al. 2009). The stability of candidate reference genes across all of the experimental conditions was analyzed using three Microsoft Excel-based software packages: geNorm (Vandesompele et al. 2002), NormFinder (Andersen et al. 2004) and BestKeeper (Pfaffl et al. 2004). All three software packages were used according to their manuals; all other multiple comparisons were performed using SPSS 17.0 (IBM, Armonk, NY, USA).
Results
Expression profiling of candidate reference genes
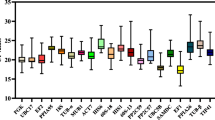
To determine the expression profiles of the candidate reference genes across all of the experimental samples, we ran an RT-qPCR, and the raw quantification cycle (C q) values were calculated. The expression levels of candidates showed a relatively wide range of expression levels and variation, as shown in the box and whisker plots (Fig. 1). MDH expression was the most abundant with the lowest mean C q value (22.68), while S24 had the lowest level of expression and its average C q value was 31.23. All genes showed expression variation among the tested samples. The lowest expression variation was observed in RPL39 with a coefficient of variation (CV) of 3.59 %, while TIP41 was the most variable across all tested samples, with a CV of 9.25 %.
Evaluation of expression stability
geNorm, NormFinder and BestKeeper were used to aid in the evaluation of suitable reference genes from the candidates. These three programs evaluate gene expression stability using distinct algorithms. The expression stability of ten candidate reference genes was evaluated in 34 cDNA samples, including roots, stems and leaves of seedlings under nitrogen starvation and PEG treatment, leaves under heat stress and flowers at different developmental stages. Altogether, the results obtained from geNorm, NormFinder, and BestKeeper were similar with regard to the best candidate reference gene for expression normalization.
geNorm analysis
The stability of potential reference genes was first examined using geNorm, in which the gene expression stability (M) of ten candidates was calculated. This program uses an M value below a threshold of 1.5 to identify reference genes with stable expression (Vandesompele et al. 2002). The lower the M value, the higher the gene expression stability. Figure 2 shows the ranking of the tested genes according to their expression stability in B. luminifera using data from four sets of treatments. When all of the samples were analyzed together, MDH and RPL39 were the most stable genes, while TIP41 and S24 were the least stable genes. In N-starved roots (NR), UBQ10 and ACT had the most stable expression levels, while in N-starved stems (NS), RPL39 and MDH had the highest expression stability. In N-starved leaves (NL), the best reference genes were MDH and EF1a. S24 was the least stable gene in all N-starved tissues tested. In the PEG-treated plants, MDH and EF1a were the most stably expressed genes in roots and stems, respectively, while EF1a and GAPDH were the most stable genes in leaves, with MDH as the next most stable. S24 and TIP41 were the most unstable genes under osmotic stress. GAPDH and TUB were the most stable genes under heat stress, while S24 and TIP41 were the least stable genes. In flowers, RPL39 and TUB were the most stable genes in the various developmental stages, while S24 and TUA showed the lowest expression stability.
To determine the optimal number of reference genes, geNorm performs a stepwise calculation of the pairwise variation (V n /V n+1) between sequential normalization factors. It is suggested that the inclusion of an additional reference gene is not required when the variation value is below 0.15 (Vandesompele et al. 2002). In our study, six of the eight experimental samples (three N-starved tissues, PEG-treated leaves, heat stress and flowering stage), V 2/3 was already below the cutoff value, suggesting that using the two most stable reference genes is sufficient for reliable gene expression normalization of target genes. While in PEG-treated roots and stems, the recommended variation value was V 5/6 and V 3/4, respectively, indicating that five and three genes are recommended for data normalization, respectively (Fig. 3).
Pairwise variation analyses of candidate reference genes in different sample sets. Pairwise variation (V n /V n+1) analyses between the normalization factors (NFn and NFn+1) were performed using the geNorm software to determine the optimal number of reference genes and carried out for qPCR data normalization in various sample pools. NR N-starved roots, NS N-starved stems, NL N-starved leaves, PR PEG treated roots, PS PEG treated stems, PL PEG treated leaves, HT heat stress treatment
NormFinder analysis
NormFinder uses a model-based approach to rank the candidate reference genes and generate a stability measure in which a lower value indicates increased stability in gene expression (Andersen et al. 2004; Zhu et al. 2013). In N-starved roots, the most stable reference genes were TUB and MDH (0.071 and 0.133, respectively) (Table 2). In the N-starved stems, TUA (0.033) and TIP41 (0.046) showed the highest expression stability. S24 was always the least stable gene. RPL39, UBQ10 and MDH were the most stable reference genes under osmotic stress, although their rankings varied slightly among tissues. These results were similar to those of geNorm. For heat stress, NormFinder ranked RPL39 and TUB as the most stable reference genes with the same stability value (0.054). UBQ10 (0.104) and S24 (0.222) were selected to be the most stable genes in flowers. Considering all treatments, MDH and RPL39 appeared most frequently among the most stable genes, while TUA and TIP41 were always the least suitable reference genes (Table 2).
BestKeeper analysis
BestKeeper determines gene expression stability of candidate reference genes based on the correlation coefficient (r), standard deviation (SD) and percentage covariance (CV) (de Carvalho et al. 2013). It is vital to emphasize that BestKeeper uses C q values instead of the raw data (relative quantities) used by geNorm and NormFinder, which may consequently lead to misinterpretation related to lack of correction. Genes with SD greater than 1 are deemed unacceptable. Those that have the lowest coefficient of variance and standard deviation (CV ± SD) are regarded as the most stable reference genes. The results of the BestKeeper analysis are shown in Table 3. In the N-starved roots, RPL39 and EF1a were identified as the most stable reference genes, TUB and TIP41 were the most stable genes in the N-starved stems, while the top ranked genes were geNorm and NormFinder. In the PEG-treated stems and leaves, MDH showed the highest expression stability. Under heat stress treatment, UBQ10 and TUB had the most stable expression levels. UBQ10 was also identified as the most stable gene in flowers. When evaluated across all the experimental samples, RPL39 and MDH ranked in the top positions in all three programs.
Reference gene evaluation
The expression patterns of SOD and NFYA-3 were evaluated using the selected reference genes. In plants, the expression of SOD is induced by heat and drought (Alscher et al. 2002; Yin et al. 2008), while transcript levels of NFYA-3 are induced by N starvation (Czechowski et al. 2005). In this study, expressions of SOD in PEG-treated and heat-stressed leaves and NFYA-3 expression in N-starved roots were assessed. The two novel recommended reference genes (RPL39 and MDH) and the most unstable gene (TIP41) were used for normalization. In all tissues tested, similar expression patterns were observed when RPL39 and MDH were used for normalization (Fig. 4). For example, in the N-starved roots, the expression level of NFYA-3 increased during continuous N-starvation, reaching a peak on day 10. In the heat-stressed leaves, the abundance of SOD also increased and reached a peak at 4 h. The expression level of SOD was increased in PEG-treated leaves for 24 h, while the abundance decreased after 5 days of continuous PEG-stress. However, when TIP41 was used for normalization, the expression patterns differed from those of RPL39 and MDH. In the N-starved roots, the transcript level of NFYA-3 peaked at 24 h and decreased thereafter. In the heat-stressed leaves, the expression level of SOD peaked at 2 h, then decreased dramatically at 4 h. The abundance of SOD was increased in PEG-treated leaves after 5 days, which differed significantly from that of RPL39 and MDH.
Expression pattern of SOD and N FYA - 3. NFYA-3 expression profile in N-starved roots (a). SOD expression profile in heat stressed leaves (b) and PEG-treated leaves (c). Normalized with two stable reference genes (RPL39 and MDH) and the most unstable reference gene (TIP41). Values are presented as mean ± SE (n = 3)
Discussion
Betula luminifera with its desirable wood properties, beautiful texture, fast growth and high adaptability to barren soil, is widely distributed in China and proposed as a priority timber species for afforestation. In addition, B. luminifera has considerable potential for use as a model tree for forest genetic studies due to its small genome (approximately 450 Mb) and comparatively short juvenile period (1.5 years) (Huang et al. 2014). Furthermore, the availability of abundant transcriptomic sequencing data and a draft genome is underway, which has encouraged many diverse molecular biological studies of B. luminifera. qPCR is widely used for accurate and rapid quantification of gene transcript levels, even for genes whose expression levels are low (Zhong et al. 2011) and is more sensitive and simple than northern blotting or semi-quantitative RT-PCR. However, the accuracy of qPCR is strongly influenced by the stability of the reference genes used for normalization. The most commonly used reference genes, such as actin and GAPDH, were chosen primarily for their known or suspected housekeeping roles in basic cellular processes, assuming that they were always stably expressed. Studies have revealed that there are no reference genes that are applicable to all plant tissues or experimental conditions (Silveira et al. 2009). Therefore, potential reference genes must be studied under different experimental treatments or for specific tissues to avoid erroneous conclusions. In this study, three algorithms (geNorm, NormFinder and BestKeeper) were used to analyze the expression stability of ten candidate genes under different abiotic stresses (N starvation, osmotic, heat) and flowering stages. To our knowledge, this is the first qPCR study of the expression stability of reference genes in B. luminifera.
Results obtained in this study show that none of the ten genes analyzed had a constant expression level in all conditions or plant tissues, which is common in plant studies (Gutierrez et al. 2008). Because these three programs are based on different algorithms and analytical procedures, they may give different results. The top stable genes (half of the total) were similar in the three algorithms for each sample set, though slight differences were found in their ranking orders. For example, in N-starved roots, geNorm selected UBQ10 as the most stable reference gene, while it was ranked third and fourth by NormFinder and BestKeeper, respectively. In N-starved leaves, GAPDH emerged as the most stable reference gene using BestKeeper, while it was ranked fourth and fifth by geNorm and NormFinder, respectively. In PEG-treated roots, TUA was identified as the most stably expressed gene by BestKeeper, while geNorm and NormFinder recommended MDH. EF1a was classified as the most stable gene in PEG-treated stems and PEG-treated leaves by BestKeeper and geNorm, respectively, but NormFinder presented different results. RPL39 and TUB were selected as the most stable reference genes in heat-stressed leaves, while they were ranked sixth and second by BestKeeper. Finally, considering all the tissues across all of treatments, RPL39 and MDH were identified as the most stably expressed genes by all three algorithms. The results of these three algorithms were considered together when determining suitable reference genes for qPCR normalization.
Among the housekeeping genes evaluated in this study, TIP41 and S24 genes were previously described as reliable reference genes in Brassica juncea across developmental stages (Chandna et al. 2012) and in coffee (Cruz et al. 2009), but they performed poorly in our experiment. Their expression levels were unstable, especially TIP41 with CV for the C q values was the highest among all samples, and both of them were transcribed at a relatively low abundance. For these reasons, we excluded these two genes from consideration as reference genes in B. luminifera. GAPDH is a traditional housekeeping gene used as a reference gene and has been validated in grapevines to be stably expressed during berry development (Park et al. 2012). In this study, GAPDH showed stable expression only in PEG-treated leaves. It displayed inappropriate expression variability, limiting its use as an internal control in Betula. TUA was not a top reference gene, although it was expressed stably under certain conditions (N-starved stems, osmotic stress). Similarly, across different tissues or under distinct hormone treatments in poplar and cucumber, TUA was the recommended gene for normalization (Alscher et al. 2002; Vandesompele et al. 2002). In contrast, TUA performed poorly across different organs, shading and girdling plus defoliation treatments in litchi (Zhong et al. 2011). TUB and UBQ10 are among the top housekeeping genes used as reference genes in plant science according to Guenin (Guenin et al. 2009). Moreover, UBQ10 was validated as suitable for normalization in Arabidopsis (Czechowski et al. 2005) and tomato (Lovdal and Lillo 2009). In Musa, TUB can be used for normalization in greenhouse leaf samples (Podevin et al. 2012). In this study, based on the results from the three programs and original C q values, UBQ10 can be considered as a stable reference gene in flowers, PEG-treated stems and PEG-treated leaves. For other treatments, fluctuations in C q values were observed. UBQ10 was suggested as inappropriate for qPCR normalization in different tissues at different developmental stages in rice (Ding et al. 2004) and soybean (Jian et al. 2008). However, in early studies of Arabidopsis and tomato, UBQ10 showed highly stable expression. In this study, results from all three software analyses showed that UBQ10 expression varied depending on the experimental condition. Consequently, it should be used with caution as an internal control. Among three potential reference genes, EF1a has been validated in cucumber (Wan et al. 2010) and strawberry (Amil-Ruiz et al. 2013) as an optimal normalization factor for gene expression under various conditions and it emerged as the most stably expressed gene in N -starved plants in our study. MDH and RPL39 are not commonly used in qPCR as reference genes, but they showed highly stable expression under almost every condition in this study. MDH was more highly ranked by geNorm under N-starvation conditions. In a previous analysis of this gene in grapevines, it did not perform well (Park et al. 2012). In contrast, similar result to ours were observed in coffee, where MDH was identified as the most stable reference gene in N starvation and heat stress (de Carvalho et al. 2013). RPL39 was ranked in the top position by the different algorithms in N starvation, osmotic and heat stresses. In melon (Kong et al. 2014) and sweet potato (Park et al. 2012), RPL was found to be a suitable reference gene. In summary, according to the results of the three statistical algorithms, RPL39, MHD and EF1a were the most stably expressed genes, and these genes can be applied to almost all treatments examined in this work, while TIP41 and S24 were the most variable. Additionally, TUB can be used as a reference gene in N-starved roots and stems and TUA can be used in N-starved leaves and stems. In PEG-treated roots and leaves, TUA and GAPDH can be used for normalization, while in PEG-treated stems and PEG-treated leaves, UB10 is a good choice. In heat stress treatments, TUB was also a good choice.
To demonstrate the need for accurate relative quantification using suitable reference genes, we determined the expression levels of SOD and NFYA-3. SOD is an important antioxidant in plants and the gene encoding SOD is induced by heat and drought stresses (Alscher et al. 2002; Yin et al. 2008). NFYA-3 is a transcription factor that responds to N starvation (Czechowski et al. 2005; Zhao et al. 2011). In this study, expression of SOD in PEG-treated and heat-stressed leaves and NFYA-3 expression in N-starved roots were assessed. Similar expression patterns were observed using the two new recommended genes (RPL39 and MDH) for normalization (Fig. 4). However, a distinctly different pattern was obtained when TIP41 was used as the reference gene. For example, when RPL39 and MDH were used for normalization, the expression level of NFYA-3 increased under continuous N-starvation, reaching a peak on day 10 in N-starved roots, whereas the transcript level of NFYA-3 peaked at 24 h and decreased significantly by day 10 using TIP41 as the reference gene. These results demonstrated that the utilization of an unsuitable reference gene could lead to inaccurate results and even opposite expression patterns. Therefore, candidate genes that are unstable under different abiotic stresses (N starvation, osmotic, heat) and flowering stages are not recommended for normalization in B. luminifera.
Conclusions
This is the first work regarding the selection of reliable reference genes for transcriptional studies in B. luminifera under N starvation, osmotic and heat stresses, and at different flower developmental stages. These results can be used as a guide for the selection of reference genes during the analysis of gene expression. This will be beneficial to future studies that require more accurate gene expression quantification in B. luminifera.
References
Alscher RG, Erturk N, Heath LS (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53:1331–1341
Amil-Ruiz F, Garrido-Gala J, Blanco-Portales R, Folta KM, Munoz-Blanco J, Caballero JL (2013) Identification and validation of reference genes for transcript normalization in strawberry (Fragaria × ananassa) defense responses. PLoS One 8(8):e70603
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Bigelow SW, Canham CD (2007) Nutrient limitation of juvenile trees in a northern hardwood forest: calcium and nitrate are preeminent. For Ecol Manag 243:310–319
Brunner AM, Yakovlev IA, Strauss SH (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4:14
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Chandna R, Augustine R, Bisht NC (2012) Evaluation of Candidate Reference Genes for Gene Expression Normalization in Brassica juncea using real time quantitative RT-PCR. PLoS One 7(5):e36918
Chen L, Zhong HY, Kuang JF, Li JG, Lu WJ, Chen JY (2011) Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234:377–390
Cruz F, Kalaoun S, Nobile P, Colombo C, Almeida J, Barros LMG, Romano E, Grossi-de-Sa MF, Vaslin M, Alves-Ferreira M (2009) Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol Breed 23:607–616
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
de Carvalho K, Bespalhok JC, dos Santos TB, de Souza SGH, Vieira LGE, Pereira LFP, Domingues DS (2013) Nitrogen starvation, salt and heat stress in coffee (Coffea arabica L.): identification and validation of new genes for qPCR normalization. Mol Biotechnol 53:315–325
Ding JY, Jia JW, Yang LT, Wen HB, Zhang CM, Liu WX, Zhang DB (2004) Validation of a rice specific gene, sucrose phosphate synthase, used as the endogenous reference gene for qualitative and real-time quantitative PCR detection of transgenes. J Agric Food Chem 52:3372–3377
Guenin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L (2009) Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot 60:487–493
Gutierrez L, Mauriat M, Guenin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6:609–618
Hong SM, Bahn SC, Lyu A, Jung HS, Ahn JH (2010) Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol 51:1694–1706
Huang HH, Jiang C, Tong ZK, Cheng LJ, Zhu MY, Lin EP (2014) Eight distinct cellulose synthase catalytic subunit genes from Betula luminifera are associated with primary and secondary cell wall biosynthesis. Cellulose 21:2183–2198
Jian B, Liu B, Bi YR, Hou WS, Wu CX, Han TF (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9:1
Kong QS, Yuan JX, Niu PH, Xie JJ, Jiang W, Huang Y, Bie ZL (2014) Screening suitable reference genes for normalization in reverse transcription quantitative real-time PCR analysis in melon. PLoS One 9(1):e87197
Lovdal T, Lillo C (2009) Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal Biochem 387:238–242
Mittler R, Blumwald E (2010) Genetic engineering for modern agriculture: challenges and perspectives. Annu Rev Plant Biol 61:443–462
Obrero A, Die JV, Roman B, Gomez P, Nadal S, Gonzalez-Verdejo CI (2011) Selection of reference genes for gene expression studies in Zucchini (Cucurbita pepo) using qPCR. J Agric Food Chem 59:5402–5411
Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10:1
Park SC, Kim YH, Ji CY, Park S, Jeong JC, Lee HS, Kwak SS (2012) Stable internal reference genes for the normalization of real-time PCR in different sweetpotato cultivars subjected to abiotic stress conditions. PLoS One 7(12):e51502
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestKeeper—excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515
Podevin N, Krauss A, Henry I, Swennen R, Remy S (2012) Selection and validation of reference genes for quantitative RT-PCR expression studies of the non-model crop Musa. Mol Breed 30:1237–1252
Silveira ED, Alves-Ferreira M, Guimaraes LA, da Silva FR, Carneiro VT (2009) Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biol 9:84
Udvardi MK, Czechowski T, Scheible WR (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20:1736–1737
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):1
Wan HJ, Zhao ZG, Qian CT, Sui YH, Malik AA, Chen JF (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399:257–261
Yin H, Chen QM, Yi MF (2008) Effects of short-term heat stress on oxidative damage and responses of antioxidant system in Lilium longiflorum. Plant Growth Regul 54:45–54
Zhao M, Ding H, Zhu JK, Zhang F, Li WX (2011) Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol 190:906–915
Zhong HY, Chen JW, Li CQ, Chen L, Wu JY, Chen JY, Lu WJ, Li JG (2011) Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep 30:641–653
Zhu J, Zhang L, Li W, Han S, Yang W, Qi L (2013) Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PLoS One 8:e53196
Author information
Authors and Affiliations
Corresponding authors
Additional information
Project funding
This work was financially supported by the National Natural Science Foundation of China (No. 31300566) and Zhejiang Province Science and Technology Support Program (No. 2012C12908-8).
The online version is available at http://www.springerlink.com
Corresponding Editor: Chai Ruihai
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, J., Zhang, J., Pan, Y. et al. Identification and evaluation of reference genes for normalization in quantitative real-time PCR analysis in the premodel tree Betula luminifera . J. For. Res. 28, 273–282 (2017). https://doi.org/10.1007/s11676-016-0314-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-016-0314-2