Abstract
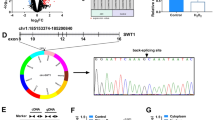
Myocardial infarction is the leading cause of death worldwide, and cardiomyocyte apoptosis during myocardial infarction and reperfusion is a significant factor of poor prognosis. As important regulatory molecules, biofunctions of circRNAs in the pathogenesis of myocardial infarction remain elusive. To confirm the expression level and biological function of circNFIX in cardiomyocytes upon oxidative stress. Divergent polymerase chain reaction and Sanger sequencing were performed to verify the circular structure. The stability of circNFIX was confirmed by RNase R treatment and actinomycin D assay. In order to simulate oxidative stress during myocardial infarction, H9c2 cells were subjected to hydrogen peroxide and hypoxia stimulation. In vivo, mouse models of myocardial ischemia were established. The biological function of circNFIX in cardiomyocytes was investigated through loss- and gain-of-function assays, and cardiomyocyte apoptosis level was detected by the terminal deoxyribonucleotidyl transferase-mediated TdT-mediated dUTP nick end labeling assay and Western blot. CircNFIX is abundant, conserved, and stable in H9c2 cells. The expression of circNFIX was significantly downregulated in cardiomyocytes subjected to oxidative stress. Enforced CircNFIX promotes H9c2 cells apoptosis induced by hydrogen peroxide, in sharp contrast to circNFIX knockdown. In this study, we found that circNFIX served as a pro-apoptosis factor in cardiomyocyte apoptosis. CircNFIX possesses potential to be the biomarker and therapeutic target in myocardial infarction.



Similar content being viewed by others
References
Aufiero S, Reckman YJ, Pinto YM, Creemers EE (2019) Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol 16:503–514
Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X (2015) The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 61:221–230
Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, Isasi CR, Jimenez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Mackey RH, Matsushita K, Mozaffarian D, Mussolino ME, Nasir K, Neumar RW, Palaniappan L, Pandey DK, Thiagarajan RR, Reeves MJ, Ritchey M, Rodriguez CJ, Roth GA, Rosamond WD, Sasson C, Towfighi A, Tsao CW, Turner MB, Virani SS, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P (2017) Heart disease and stroke Statistics-2017 update: a report from the American Heart Association. Circulation 135:e146–e603
Biressi S, Tagliafico E, Lamorte G, Monteverde S, Tenedini E, Roncaglia E, Ferrari S, Ferrari S, Cusella-De Angelis MG, Tajbakhsh S, Cossu G (2007) Intrinsic phenotypic diversity of embryonic and fetal myoblasts is revealed by genome-wide gene expression analysis on purified cells. Dev Biol 304:633–651
Cai L, Qi B, Wu X, Peng S, Zhou G, Wei Y, Xu J, Chen S, Liu S (2019) Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J Mol Cell Cardiol 130:10–22
Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, Garzia L, Torchia J, Nor C, Morrissy AS, Agnihotri S, Thompson YY, Kuzan-Fischer CM, Farooq H, Isaev K, Daniels C, Cho BK, Kim SK, Wang KC, Lee JY, Grajkowska WA, Perek-Polnik M, Vasiljevic A, Faure-Conter C, Jouvet A, Giannini C, Nageswara Rao AA, Li KKW, Ng HK, Eberhart CG, Pollack IF, Hamilton RL, Gillespie GY, Olson JM, Leary S, Weiss WA, Lach B, Chambless LB, Thompson RC, Cooper MK, Vibhakar R, Hauser P, van Veelen MC, Kros JM, French PJ, Ra YS, Kumabe T, López-Aguilar E, Zitterbart K, Sterba J, Finocchiaro G, Massimino M, Van Meir EG, Osuka S, Shofuda T, Klekner A, Zollo M, Leonard JR, Rubin JB, Jabado N, Albrecht S, Mora J, Van Meter TE, Jung S, Moore AS, Hallahan AR, Chan JA, Tirapelli DPC, Carlotti CG, Fouladi M, Pimentel J, Faria CC, Saad AG, Massimi L, Liau LM, Wheeler H, Nakamura H, Elbabaa SK, Perezpeña-Diazconti M, Chico Ponce de León F, Robinson S, Zapotocky M, Lassaletta A, Huang A, Hawkins CE, Tabori U, Bouffet E, Bartels U, Dirks PB, Rutka JT, Bader GD, Reimand J, Goldenberg A, Ramaswamy V, Taylor MD 2017. Intertumoral heterogeneity within medulloblastoma subgroups Cancer Cell, 31: 737-54.e6
Chaudhry AZ, Lyons GE, Gronostajski RM (1997) Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev Dyn 208:313–325
Chen G, Shi Y, Liu M, Sun J (2018) circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis 9:175
Cui X, Wang J, Guo Z, Li M, Li M, Liu S, Liu H, Li W, Yin X, Tao J, Xu W (2018) Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer 17:123
Dixon C, Harvey TJ, Smith AG, Gronostajski RM, Bailey TL, Piper M (2013) Nuclear factor one X regulates Bobby sox during development of the mouse forebrain. Cell Mol Neurobiol 33:867–873
Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB (2017a) Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 38:1402–1412
Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB (2017b) Identifying and characterizing circRNA-protein interaction. Theranostics 7:4183–4191
Floris G, Zhang L, Follesa P, Sun T (2017) Regulatory role of circular RNAs and neurological disorders. Mol Neurobiol 54:5156–5165
Fraser J, Essebier A, Gronostajski RM, Boden M, Wainwright BJ, Harvey TJ, Piper M (2017) Cell-type-specific expression of NFIX in the developing and adult cerebellum. Brain Struct Funct 222:2251–2270
Glazar P, Papavasileiou P, Rajewsky N (2014) circBase: a database for circular RNAs. Rna 20:1666–1670
Hall T, Walker M, Ganuza M, Holmfeldt P, Bordas M, Kang G, Bi W, Palmer LE, Finkelstein D, McKinney-Freeman S (2018) Nfix promotes survival of immature hematopoietic cells via regulation of c-Mpl. Stem Cells 36:943–950
Han B, Chao J, Yao H (2018) Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther 187:31–44
Hausenloy DJ, Duchen MR, Yellon DM (2003) Inhibiting mitochondrial permeability transition pore opening at reperfusion protects against ischaemia-reperfusion injury. Cardiovasc Res 60:617–625
Heng YH, McLeay RC, Harvey TJ, Smith AG, Barry G, Cato K, Plachez C, Little E, Mason S, Dixon C, Gronostajski RM, Bailey TL, Richards LJ, Piper M (2014) NFIX regulates neural progenitor cell differentiation during hippocampal morphogenesis. Cereb Cortex 24:261–279
Holdt LM, Kohlmaier A, Teupser D (2018) Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci 75:1071–1098
Huang S, Li X, Zheng H, Si X, Li B, Wei G, Li C, Chen Y, Chen Y, Liao W, Liao Y, Bin J (2019) Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation 139:2857–2876
Jafari Ghods F (2018) Circular RNA in saliva. Adv Exp Med Biol 1087:131–139
Li M, Ding W, Tariq MA, Chang W, Zhang X, Xu W, Hou L, Wang Y, Wang J (2018a) A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics 8:5855–5869
Li X, Yang L, Chen LL (2018b) The biogenesis, functions, and challenges of circular RNAs. Mol Cell 71:428–442
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G (2015a) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22:256–264
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Jianren G, He X, Huang S (2015b) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25:981–984
Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM, Zhang JF (2019) Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol 20:84
Lim TB, Aliwarga E, Luu TDA, Li YP, Ng SL, Annadoray L, Sian S, Ackers-Johnson MA, Foo RS (2019) Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc Res 115:1998–2007
Mao Y, Liu J, Zhang D, Li B (2015) MiR-1290 promotes cancer progression by targeting nuclear factor I/X(NFIX) in esophageal squamous cell carcinoma (ESCC). Biomed Pharmacother 76:82–93
Matuzelski E, Bunt J, Harkins D, Lim JWC, Gronostajski RM, Richards LJ, Harris L, Piper M (2017) Transcriptional regulation of Nfix by NFIB drives astrocytic maturation within the developing spinal cord. Dev Biol 432:286–297
O'Connor C, Campos J, Osinski JM, Gronostajski RM, Michie AM, Keeshan K (2015) Nfix expression critically modulates early B lymphopoiesis and myelopoiesis. PLoS One 10:e0120102
Piper M, Gronostajski R, Messina G (2019) Nuclear factor one X in development and disease. Trends Cell Biol 29:20–30
Riddell J, Gazit R, Garrison BS, Guo G, Saadatpour A, Mandal PK, Ebina W, Volchkov P, Yuan GC, Orkin SH, Rossi DJ (2014) Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell 157:549–564
Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, Tang W, Cao H (2017) An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 8:73271–73281
Rossi G, Antonini S, Bonfanti C, Monteverde S, Vezzali C, Tajbakhsh S, Cossu G, Messina G (2016) Nfix regulates temporal progression of muscle regeneration through modulation of Myostatin expression. Cell Rep 14:2238–2249
Stringer BW, Bunt J, Day BW, Barry G, Jamieson PR, Ensbey KS, Bruce ZC, Goasdoué K, Vidal H, Charmsaz S, Smith FM, Cooper LT, Piper M, Boyd AW, Richards LJ (2016) Nuclear factor one B (NFIB) encodes a subtype-specific tumour suppressor in glioblastoma. Oncotarget 7:29306–29320
Sun H, Tang W, Rong D, Jin H, Fu K, Zhang W, Liu Z, Cao H, Cao X (2018) Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark 21:299–306
Tan WL, Lim BT, Anene-Nzelu CG, Ackers-Johnson M, Dashi A, See K, Tiang Z, Lee DP, Chua WW, Luu TD, Li PY, Richards AM, Foo RS (2017) A landscape of circular RNA expression in the human heart. Cardiovasc Res 113:298–309
Townsend N, Wilson L, Bhatnagar P, Wickramasinghe K, Rayner M, Nichols M (2016) Cardiovascular disease in Europe: epidemiological update 2016. Eur Heart J 37:3232–3245
Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP, Li YR, Li PF (2011) miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med 17:71–78
Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, Chen C, Yan KW, Ponnusamy M, Zhang YH, Li PF (2017) Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ 24:1111–1120
Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF (2016) A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 37:2602–2611
Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, Fan YY, Li PF (2014) CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun 5:3596
Wang K, Zhou LY, Wang JX, Wang Y, Sun T, Zhao B, Yang YJ, An T, Long B, Li N, Liu CY, Gong Y, Gao JN, Dong YH, Zhang J, Li PF (2015) E2F1-dependent miR-421 regulates mitochondrial fragmentation and myocardial infarction by targeting Pink1. Nat Commun 6:7619
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, Wang G, Wu P, Wang H, Jiang L, Yuan W, Sun Z, Ming L (2019) Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer 18:116
Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S (2016) Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol 98:103–107
Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, Xiong F, Guo C, Wu X, Li Y, Li X, Li G, Zeng Z, Xiong W (2020) Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer 19:22
Xu H, Zhang Y, Qi L, Ding L, Jiang H, Yu H (2018) NFIX circular RNA promotes glioma progression by regulating miR-34a-5p via notch signaling pathway. Front Mol Neurosci 11:225
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, Huang S, Xie B, Zhang N (2018) Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst 110:304–315
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B, Wang S (2018) CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis 9:417
Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, Yang W, Zhang C, Yang Q, Yee A, Chen Y, Yang F, Sun H, Huang R, Yee AJ, Li RK, Wu Z, Backx PH, Yang BB (2017) A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 7:3842–3855
Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, Li Z, Ming L, Xie B, Zhang N (2018a) A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37:1805–1814
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, Liu H, Xu J, Xiao F, Zhou H, Yang X, Huang N, Liu J, He K, Xie K, Zhang G, Huang S, Zhang N (2018b) A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 9:4475
Zhang S-J, Xue C, Li C-P, Li X-M, Liu C, Liu B-H, Shan K, Jiang Q, Zhao C, Yan B (2017) Identification and characterization of circular RNAs as a new class of putative biomarkers in diabetes retinopathy. Invest Ophthalmol Vis Sci 58:6500–6509
Funding
The support was provided by the National Natural Science Foundation of China (81770900), the Science and Technology Development Foundation of Shandong Province (2014GHY115025).
Author information
Authors and Affiliations
Contributions
X. C. and Y. D. contributed equally to this study. W. X., X. C., and Y. D. contributed to the design of this study. X. C., Y. D., X. W., M. J., W. Y., G. L., and S. S. performed all the experiments. X. C. and M. L. performed the data collection and analysis. X. C. and Y. D. contributed to the interpretation of data and drafting the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Animal studies
All institutional and national guidelines for the care and use of laboratory animals were followed and approved by the ethical standards of the Animal Care Committee, Qingdao University.
Human subjects/informed consent statement
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Editor: Tetsuji Okamoto
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 373 kb)
Rights and permissions
About this article
Cite this article
Cui, X., Dong, Y., Li, M. et al. A circular RNA from NFIX facilitates oxidative stress-induced H9c2 cells apoptosis. In Vitro Cell.Dev.Biol.-Animal 56, 715–722 (2020). https://doi.org/10.1007/s11626-020-00476-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-020-00476-z