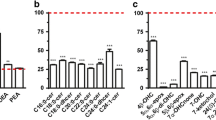
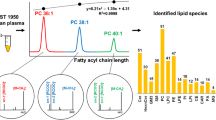
Abstract
Objective
Cholesteryl esters (CEs) are composed of various fatty acyl chains attached to the hydroxyl groups of cholesterol, and abnormalities in their metabolism are related to many diseases. This study aimed to develop an ultrahigh-performance liquid chromatography-quadrupole exactive mass spectrometry (UPLC-Q-Exactive MS) method to identify the CEs in plasma.
Methods
First, the MS fragmentation patterns were investigated using seven commercial CE standards. Then, the CEs in plasma were characterized through the accurate mass data of precursor ions and characteristic product ions. A strategy of step-by-step m/z scans in a narrow range was proposed to identify more trace CEs by the full-scan data-dependent MS/MS (ddMS2) mode.
Results
A total of 50 CE species consisting of 55 regioisomers were identified in human plasma. Among them, two species were reported for the first time.
Conclusion
This study is the most comprehensive identification of CE species in human plasma to date. These results will contribute to the in-depth profiling of CEs in human plasma and provide guidance for animal model selection when studying lipid-related diseases.
Similar content being viewed by others
References
Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol, 2020,21(4):225–245
Yang W, Bai Y, Xiong Y, et al. Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature, 2016,531(7596):651–655
Shibuya Y, Chang CCY, Chang TY. ACAT1/SOAT1 as a therapeutic target for Alzheimer’s disease. Future Med Chem, 2015,7(18):2451–2467
Garcia-Bermudez J, Birsoy K. Drugging ACAT1 for Cancer Therapy. Mol Cell, 2016,64(5):856–857
Quehenberger O, Armando AM, Brown AH, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res, 2010,51(11):3299–3305
Talbot CPJ, Plat J, Joris PJ, et al. HDL cholesterol efflux capacity and cholesteryl ester transfer are associated with body mass, but are not changed by diet-induced weight loss: A randomized trial in abdominally obese men. Atheroscler, 2018,274(1):23–28
Gerl MJ, Vaz WLC, Domingues N, et al. Cholesterol is Inefficiently Converted to Cholesteryl Esters in the Blood of Cardiovascular Disease Patients. Sci Rep, 2018,8(1):14764
Sekiya M, Osuga JI, Nagashima S, et al. Ablation of Neutral Cholesterol Ester Hydrolase 1 Accelerates Atherosclerosis. Cell Metab, 2009,10(3):219–228
Proitsi P, Kim M, Whiley L, et al. Plasma lipidomics analysis finds long chain cholesteryl esters to be associated with Alzheimer’s disease. Transl Psychiatry, 2015,5(1):e494
Mayengbam SS, Singh A, Pillai AD, et al. Influence of cholesterol on cancer progression and therapy. Transl Oncol, 2021,14(6):101043
Prada M, Wittenbecher C, Eichelmann F, et al. Association of the odd-chain fatty acid content in lipid groups with type 2 diabetes risk: A targeted analysis of lipidomics data in the EPIC-Potsdam cohort. Clin Nutr, 2021,40(8):4988–4999
Zhou J, Chen X, Chen W, et al. Comprehensive plasma metabolomic and lipidomic analyses reveal potential biomarkers for heart failure. Mol Cell Biochem, 2021,476(9):3449–3460
Liu T, Peng F, Yu J, et al. LC-MS-based lipid profile in colorectal cancer patients: TAGs are the main disturbed lipid markers of colorectal cancer progression. Anal Bioanal Chem, 2019,411(20):5079–5088
Tarasov K, Ekroos K, Suoniemi M, et al. Molecular lipids identify cardiovascular risk and are efficiently lowered by simvastatin and PCSK9 deficiency. J Clin Endocrinol Metab, 2014,99(1):E45–52
Afshinnia F, Rajendiran TM, Karnovsky A, et al. Lipidomic Signature of Progression of Chronic Kidney Disease in the Chronic Renal Insufficiency Cohort. Kidney Int Rep, 2016,1(4):256–268
Chan RB, Oliveira TG, Cortes EP, et al. Comparative Lipidomic Analysis of Mouse and Human Brain with Alzheimer Disease. J Biol Chem, 2012,287(4):2678–2688
Michalec C, Sulc M, Mestan J. Analysis of cholesteryl esters and triglycerides by thin-layer chromatography. Nature, 1962,193(4810):63–64
Subramanian A, Joshi BS, Roy AD, et al. NMR spectroscopic identification of cholesterol esters, plasmalogen and phenolic glycolipids as fingerprint markers of human intracranial tuberculomas. NMR Biomed, 2008,21(3):272–288
Asmis R, Buhler E, Jelk J, et al. Concurrent quantification of cellular cholesterol, cholesteryl esters and triglycerides in small biological samples — Reevaluation of thin layer chromatography using laser densitometry. J Chromatogr B, 1997,691(1):59–66
Hammann S, Vetter W. Gas chromatographic separation of fatty acid esters of cholesterol and phytosterols on an ionic liquid capillary column. J Chromatogr B, 2015,1007(1):67–71
Liebisch G, Binder M, Schifferer R, et al. High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS). Biochim Biophys Acta Mol Cell Biol Lipids, 2006,1761(1):121–128
Lin M, Wang Z, Wang D, et al. Mathematical Model-Assisted UHPLC-MS/MS Method for Global Profiling and Quantification of Cholesteryl Esters in Hyperlipidemic Golden Hamsters. Anal Chem, 2019, 91(7):4504–4512
Triebl A, Troetzmueller M, Hartler J, et al. Lipidomics by ultrahigh performance liquid chromatography-high resolution mass spectrometry and its application to complex biological samples. J Chromatogr B, 2017, 1053(1):72–80
Sala P, Poetz S, Brunner M, et al. Determination of Oxidized Phosphatidylcholines by Hydrophilic Interaction Liquid Chromatography Coupled to Fourier Transform Mass Spectrometry. Int J Mol Sci, 2015,16(4):8351–8363
Contrepois K, Mahmoudi S, Ubhi BK, et al. Cross-Platform Comparison of Untargeted and Targeted Lipidomics Approaches on Aging Mouse Plasma. Sci Rep, 2018,8(1):17747
Cajka T, Smilowitz JT, Fiehn O. Validating Quantitative Untargeted Lipidomics Across Nine Liquid Chromatography-High-Resolution Mass Spectrometry Platforms. Anal Chem, 2017,89(22):12360–12368
Koelmel JP, Kroeger NM, Gill EL, et al. Expanding Lipidome Coverage Using LC-MS/MS Data-Dependent Acquisition with Automated Exclusion List Generation. J Am Soc Mass Spectrom, 2017,28(5):908–917
Zhou J, Li Y, Chen X, et al. Development of data-independent acquisition workflows for metabolomic analysis on a quadrupole-orbitrap platform. Talanta, 2017,164(1):128–136
Matyash V, Liebisch G, Kurzchalia TV, et al. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res, 2008,49(5):1137–1146
Hasan M, Siegmund W, Oswald S. Rapid LC-MS/MS method for the determination of 4-hydroxycholesterol/cholesterol ratio in serum as endogenous biomarker for CYP3A activity in human and foals. J Chromatogr B, 2016,1033(1):193–199
Yu S, Dong J, Zhou W, et al. A rapid and precise method for quantification of fatty acids in human serum cholesteryl esters by liquid chromatography and tandem mass spectrometry. J Chromatogr B, 2014,960(1):222–229
Cajka T, Fiehn O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Anal Chem, 2014,61(1):192–206
Gardner MS, McWilliams LG, Jones JI, et al. Simultaneous Quantification of Free Cholesterol, Cholesteryl Esters, and Triglycerides without Ester Hydrolysis by UHPLC Separation and In-Source Collision Induced Dissociation Coupled MS/MS. J Am Soc Mass Spectrom, 2017,28(11):2319–2329
Yan Z, Yan R. Improved Data-Dependent Acquisition for Untargeted Metabolomics Using Gas-Phase Fractionation with Staggered Mass Range. Anal Chem, 2015,87(5):2861–2868
Fenaille F, Barbier Saint-Hilaire P, Rousseau K, et al. Data acquisition workflows in liquid chromatography coupled to high resolution mass spectrometry-based metabolomics: Where do we stand? J Chromatogr A, 2017,1526(1):1–12
Hu B, Song C, Li L, et al. Qualitative distribution of endogenous phosphatidylcholine and sphingomyelin in serum using LC-MS/MS based profiling. J Chromatogr B, 2020,1155(1):122289
Liu X, Wang J, Hu B, et al. Qualitative distribution of endogenous sphingolipids in plasma of human and rodent species by UPLC-Q-Exactive-MS. J Chromatogr B, 2021,1173(1):122684
Acknowledgments
The authors thank Hongren Biopharmaceutical Inc., Wuhan, China, for providing technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
This work was supported by National Natural Science Foundation of China (No. 81874309, No. 81903901 and No. 81803717) and the Youth Foundation of the Zhejiang Academy of Medical Sciences (No. 2020Y003).
Supplementary data
Rights and permissions
About this article
Cite this article
Wang, Jc., Liu, Xc., Cao, P. et al. Qualitative Distribution of Endogenous Cholesteryl Esters in Plasma of Humans and Three Rodent Species Using Stepwise UPLC-Q-Exactive-MS. CURR MED SCI 42, 692–701 (2022). https://doi.org/10.1007/s11596-022-2577-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11596-022-2577-5