Abstract
In this study, we investigated two unidentified lineages of root-colonizing fungi belonging to the order Pleosporales (Dothideomycetes), which were isolated from Festuca vaginata (Poaceae), a dominant grass species in the semiarid sandy grasslands of Hungary. For molecular phylogenetic studies, seven loci (internal transcribed spacer, partial large subunit and small subunit region of nrRNA, partial transcription elongation factor 1-α, RNA polymerase II largest subunit, RNA polymerase II second largest subunit, and ß-tubulin genes) were amplified and sequenced. Based on morphology and multilocus phylogenetic analyses, we found that one lineage belonged to Delitschia chaetomioides P. Karst. (Delitschiaceae), and the isolates of the other lineage represented a novel monotypic genus in the family Trematosphaeriaceae (suborder Massarineae). For this lineage, we proposed a new genus, Fuscosphaeria, represented by a single species, F. hungarica. In both lineages, only immature and degenerated sporocarps could be induced. These were sterile, black, globose, or depressed globose structures with numerous mycelioid appendages submerged in culture media or on the surface of autoclaved plant materials. Both species are first reported here as root-colonizing fungi.
Similar content being viewed by others
Introduction
Fungal root endophytes are common non-pathogenic colonizers of various plant species, including gramineous hosts and woody species (Rodriguez et al. 2009; Sieber and Grünig 2013; Lukešová et al. 2015). This artificial group of fungi also comprises pathogens and saprobes, which, at some point in their life cycle, colonize plant tissues without causing symptoms of tissue damage (Wilson 1995); and it seems that in this group, species commonly considered as endophytes are rather pathogenic (Schlegel et al. 2016) or possess an expanded repertoire of genes linked with saprobic abilities (Knapp et al. 2018). These root colonizers are relatively frequent in arid and semiarid grasslands worldwide (Mandyam and Jumpponen 2005), where they inhabit healthy roots of different grass species (Porras-Alfaro et al. 2008; Knapp et al. 2012, 2019). They are generally ascomycetes and usually have pigmented hyphae, which is why they are commonly called dark septate endophytes (DSE) (Jumpponen and Trappe 1998; Porras-Alfaro and Bayman 2011). The root-associated fungal communities of gramineous plants in grassland ecosystems are diverse (e.g., Khidir et al. 2010; Li et al. 2018). Festuca vaginata Waldst. et Kit. ex Willd. is one of the dominant grass species in sandy areas, such as the semiarid open sandy grasslands of the interfluves of the Danube and Tiscia (Szabó et al. 2017), which are also inhabited by a wide spectrum of root-colonizing non-mycorrhizal fungi (Knapp et al. 2012, 2015) as well as arbuscular mycorrhizae (Endresz et al. 2013). Similar to other grasses dominating certain ecosystems across the temperate region, such as Andropogon, Bouteloua, Festuca, and Stipa species (e.g., Porras-Alfaro et al. 2008; Mandyam et al. 2010; Knapp et al. 2012, 2019) F. vaginata accommodates a core community of root-associated fungi, which was concluded using isolation-based techniques (Khidir et al. 2010; Knapp et al. 2012, 2019).
The order Pleosporales is one of the most common orders in grassland ecosystems, comprising a plethora of grass root endophytes (Zhang et al. 2012; Jumpponen et al. 2017). Several common pleosporalean DSE fungi have been studied to date, including Darksidea species (Knapp et al. 2015, 2019) and the relatively well-studied Periconia macrospinosa (see Mandyam et al. 2010; Knapp et al. 2018). Pleosporales includes an increasing number of root endophytic species and genera; for example, in the last year only, several novel DSE lineages were investigated and formally described, such as Alfoldia vorosii (Crous et al. 2019), Laburnicola rhizohalophila (Yuan et al. 2019), Kiskunsagia ubrizsyi (Crous et al. 2019), and Posidoniomyces atricolor (Vohník et al. 2019). Pleosporalean root endophytes belong to various families, some within the suborders Pleosporineae and Massarineae. To date, root-colonizing fungi have not been reported in the families Delitschiaceae and Trematosphaeriaceae.
The family Delitschiaceae is a basal family (along with Massariaceae) in the order Pleosporales (Zhang et al. 2012; Hyde et al. 2013), comprising mainly terrestrial and saprobic species occurring on herbivore dung (e.g., Kruys et al. 2006; Doveri 2011), aged wood, and plants (Hyde et al. 2013) or submerged wood (Hyde and Steinke 1996; Rivera-Chávez et al. 2019); one species was identified from decaying fruits of Nypa fruticans (Jayasiri et al. 2019). The family is monogeneric, including only the genus Delitschia, with relatively few species (Jayasiri et al. 2019). Similarly to Delitschia species, species in the family Trematosphaeriaceae (suborder Massarineae) are usually saprobic fungi living on decomposed woody materials submerged in mangroves, and they were also recorded on the oil palm, which is a terrestrial plant species (Suetrong et al. 2011). In addition, pathogenic species from the family Trematosphaeriaceae were also reported as the causal agents of human mycetoma (Ahmed et al. 2014). This family includes three genera, Falciformispora, Halomassarina, and Trematosphaeria (Suetrong et al. 2011).
During our investigations of root-colonizing fungi of dominant grass species in the semiarid sandy open grasslands of Kiskunság in the Great Hungarian Plain, we isolated some fungi from healthy roots of F. vaginata, and found that they represented two distinct taxa. The present study aimed to identify these taxa based on multilocus molecular phylogeny and morphological comparison of the obtained isolates.
Materials and methods
Plant material sampling and fungi isolation
The roots of Festuca vaginata were sampled in the semiarid grassland of the Great Hungarian Plain near Fülöpháza, Hungary (N46° 52′, E19° 25′) during the spring of 2014 (for detailed description of the study site, see Kovács and Szigetvári 2002 and Knapp et al. 2012). Isolates were collected from surface-sterilized healthy roots as described in Knapp et al. (2012). We selected ten isolates representing two different lineages (DSE-87 and DSE-88 groups, with five isolates per group) (Table 1). The holotype specimen of the novel taxon was dried, and in form of a biologically inert agar culture, it was deposited in the herbarium of the Hungarian Natural History Museum, Budapest (BP) under the accession number 111139BP. Ex-type and other cultures investigated in this study were deposited in the culture collection of the Westerdijk Fungal Biodiversity Institute (CBS 147250–147251), and nomenclatural novelties and descriptions were deposited in MycoBank (www.MycoBank.org, Crous et al. 2004).
Fungal morphology and sporulation
The five isolates of each taxon were subcultured onto PDA and MEA media inoculated with fungal plugs (5 mm diameter) in Petri dishes (5 cm diameter). Growth rate and colony characteristics were recorded in cultures grown for 2 or 5 weeks at 22 °C. Morphological characteristics of the fungal structures were examined by bright-field and phase-contrast microscopy using a ZEISS AxioScope2 microscope equipped with an AxioCam ICc5 camera (Zeiss, Germany) along with the Zeiss ZEN 2011 software. Measurements and photographs were made using structures mounted in clear lactic acid. Colors were designated according to Rayner (1970).
To induce sporulation, the isolates were cultured on autoclaved pine needles and stinging nettle stems laid on water agar (WA) media in Petri dishes (9 cm diameter) at 22 °C for 3 months. The isolates were also cultured on WA media supplemented with minced vegetables (carrot, turnip, celery, and kohlrabi) at a pH of 3.5.
DNA extraction and amplification
Genomic DNA was extracted from fungal mycelia using the DNeasy Plant Mini Kit (Qiagen) following the manufacturer’s instructions. Seven loci were amplified and sequenced: internal transcribed spacer (ITS), partial 18S small subunit (SSU) and partial 28S large subunit (LSU) of the nrDNA, partial beta-tubulin gene (TUB), translation elongation factor 1-alpha gene (TEF), partial RNA Polymerase II largest subunit gene (RPB1), and partial RNA polymerase II second largest subunit gene (RPB2). The following primers were used for the amplification and sequencing: for ITS, ITS1F/ITS4 (White et al. 1990; Gardes and Bruns 1993); for SSU, NS1/NS4 (White et al. 1990); for LSU, LROR/LR6 (Rehner and Samuels 1994; Vilgalys and Hester 1990); for TUB, Bt2a/Bt2b (Glass and Donaldson 1995); for TEF, EF1-983/EF1-2218R (Rehner and Buckley 2005); for RPB1, RPB1-Af/RPB1-Cr (Stiller and Hall 1997; Matheny et al. 2002); and for RPB2, RPB2-6F/RPB2-7R (Liu et al. 1999). Additional primers, D2for (ATAAAACGGCCGTGACTGTC) and D2rev (GAGGGGATTAAGAGATCC), were used for sequencing the ITS region of DSE-87 isolates. The RPB1 and TUB and regions of some isolates were sequenced but were not used in the phylogenetic analyses (Table 1).
All PCR amplifications were performed in a final volume of 20 μL. Reaction components included 1 μL of 10 μM forward and reverse primers (Sigma-Aldrich, Germany), 2 μL of the DNA template, and 10 μL of the Dream Taq Green PCR Master Mix (Thermo Fisher Scientific, USA), or Phusion Green Hot Start II High-Fidelity PCR Master Mix (Thermo Fisher Scientific, USA) for the ITS of isolates of DSE-87 and RPB1 of the DSE-88 group.
The cycling times and temperatures were as follows: an initial denaturation step for 10 min at 95 °C; 35 cycles at 95 °C for 45 s (for ITS and RPB2), 30 s (for LSU and TEF), or 15 s (for SSU and RPB2); 1 min (for SSU, LSU, and TEF), 45 s (for ITS), or 30 s (RPB2) at an annealing temperature of 55 °C (for ITS and TEF), 52 °C (for SSU and RPB2), or 48 °C (foe LSU); 3 min (for TEF), 90 s (SSU and LSU), or 1 min (for RPB2 and ITS) at 72 °C; and a final extension step of 10 min (for ITS) or 7 min (for SSU, LSU, TEF, and RPB2). Additionally, RPB1 amplifications were performed with the following parameters: 98° °C for 2 min, followed by 15 cycles of 10 s at 98 °C, 20 s at 67 °C and 13 s at 72 °C, and 30 cycles of 10 s at 98 °C, 20 s at 52 °C, and 15 s at 72 °C, with a final extension step of 2 min at 72 °C. Additionally, for the ITS of the DSE-87 group, cycling times and temperatures were as follows: 98 °C for 2 min, followed by 36 cycles of 5 s at 98 °C, 5 s at 60 °C, and 15 s at 72 °C, and a final extension step of 5 min at 72 °C. The PCR products were separated on 1.5% agarose gel containing GelRed (Biotium Inc., CA, USA) in 0.5 × TBE buffer and visualized under UV light.
The sequences were compiled from electropherograms using the Pregap4 and Gap4 software packages (Staden et al. 2000) and deposited in GenBank (Table 1). The obtained sequences were compared with the accessions in the National Center for Biotechnology Information database (NCBI, http://www.ncbi.nlm.nih.gov/Blast.cgi) using the BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al. 1990).
Phylogenetic analyses
We combined and aligned our sequences with those from representative taxa in GenBank using the online version of MAFFT 7 (Katoh and Standley 2013) and the E-INS-i method. The alignments were examined and edited using MEGA 7 (Kumar et al. 2016). In the case of the DSE-87 group, a family level dataset was used to obtain information about the phylogenetic position of our sequences among all sequences and taxa of Delitschiaceae and representative sequences from Massariaceae and other related families (Supplementary Table 1). In the phylogenetic analysis, Hysterium angustatum (CBS 123334) and Psiloglonium araucanum (CBS 112412) served as multiple outgroups. In this dataset, for multilocus analysis, we used ITS, LSU, SSU, TEF, and RPB2 as well as the indels coded from the ITS, LSU, and SSU regions (Nagy et al. 2012) using a simple indel coding algorithm (Simmons et al. 2001; Young and Healy 2003) with the program FASTGAP (Borchsenius 2009). Therefore, in the first dataset, eight partitions were used. The second dataset contained sequences of DSE-88 group and taxa representing lineages of the pleosporalean suborder Massarineae (Supplementary Table 2), in which they grouped. In this analysis, ITS, LSU, SSU, and TEF were used as four partitions and Leptosphaeria doliolum var. doliolum (CBS 505.75) served as outgroup. Bayesian inference (BI) analyses were performed with MRBAYES 3.1.2 (Ronquist and Huelsenbeck 2003) using a GTR + G substitution model for the nucleotide partitions and the two-parameter Markov (Mk2 Lewis) model for the indel partition. Four Markov chains were run for 10,000,000 generations sampling every 1000 generations with a burn-in value set at 4000 sampled trees. Maximum likelihood (ML) phylogenetic analysis was carried out with the RAXMLGUI 1.3 implementation (Silvestro and Michalak 2012; Stamatakis 2014). A GTR + G nucleotide substitution model was used for nucleotide partitions with ML estimation of base frequencies, and the indel data were treated as binary data. ML bootstrap (BS) analysis with 1000 replicates was used to test the support of the branches. Phylogenetic trees were visualized and edited in MEGA 7 (Kumar et al. 2016) and deposited in TreeBASE (www.treebase.org) as study S26261.
Results
Morphology
The colonies of DSE-87 isolates on MEA were brownish grey, fluffy with an abundant aerial mycelium, and with pale brown or white marginal zone. PDA colonies were olivaceous-grey or greenish white, fluffy with a sparse aerial mycelium, and with brownish white marginal zone. Terminal and intercalary chlamydospores were formed (Fig. 1), and sporocarp-like structures could be produced. These were sterile, black, and globose morphs with numerous mycelioid appendages, which were observed when the isolates were cultured on the WA media supplemented with minced vegetables or pine needles and kept at 20 °C 3 months after inoculation (Fig. 1). Discoloration (color change from orange to white) of the WA media with minced vegetables was noticed.
The colonies of DSE-88 isolates on the three different media were yellowish to dark grey, flat, and they changed the agar color from orange or light brown to dark brown (Fig. 2). In addition to chlamydospores (Fig. 2), sterile, depressed, and globose sporocarp-like structures were observed on the surface of autoclaved stinging nettles and on isolates cultured on the WA media supplemented with minced vegetables or pine needles (for detailed colony description, see the taxonomy section).
Fuscosphaeria hungarica (ex-holotype, CBS 147250). a Colony on MEA. b Colony on PDA. c, d Sporocarp-like structure produced on the surface of stinging nettle. e Developing sporocarp-like structure. f Cross section of a sporocarp-like structure formed submerged in WA media supplemented with pine needles. g Terminal chlamydospores produced on PDA. Scale bars: 50 μm
Neither mature sexual or asexual morphs nor ascospore or conidium production by the strains was observed in any of the media or conditions tested, or in the laboratory where the strains were maintained and kept for years before the present study.
Molecular phylogeny
The targeted DNA sequences of the isolates could be used for the phylogenetic analyses (ITS, LSU, SSU, and TEF for both groups, and RPB2 for the DSE-87 group), excluding the TEF region of strain DSE874 and RPB2 region of strain DSE872, which failed to be amplified (Table 1). Sequencing of the ITS region of the DSE-87 isolates was problematic because amplification of this region resulted in an unusually large (~ 2200 bp) amplicon/product. Thus, for sequencing the whole ITS region, two additional primers (D2for and D2rev) were needed.
Based on the BLAST search of the NCBI’s GenBank nucleotide database, the closest ITS hits of the DSE-87 isolates showed no unambiguous relations to a certain species; only weak similarities with sequences of certain pleosporalean species were observed. Using the LSU, SSU, and TEF sequences, relatively clear similarities to a Delitschia species were found (Supplementary Table 3). Results of BLAST analyses of different DSE-88 isolate sequences showed no very similar hits (Supplementary Table 4).
For the DSE-87 group, the combined dataset consisted of 32 taxa and 7965 characters, and the group was positioned together with the strains of Delitschia chaetomioides P. Karst. within the family Delitschiaceae (Fig. 3). The five isolates formed a well-supported (PP = 1, ML BS = 99) distinct clade besides four D. chaetomioides strains. These nine strains together formed strongly supported (PP = 1, ML BS = 90) clades with strong internal clades, showing the unambiguous position of the isolates presented here as D. chaetomioides (Fig. 3).
Phylogenetic tree of Delitschia species and related representative taxa from basal groups of Pleosporales. Strains presented in this study are shown in bold. The 50% majority rule consensus phylogram inferred from the Bayesian analysis of the combined dataset of five loci (ITS, LSU, SSU, TEF, RPB2) and coded indels from ITS, LSU, and SSU as three additional partitions. Bayesian posterior probabilities (≥ 90) are shown before slashes, ML bootstrap support values (≥ 70) are shown after slashes. Hysterium angustatum (CBS 123334) and Psiloglonium araucanum (CBS 112412) served as multiple outgroups. Highlighted sections indicate affiliations to species or families. The scale bar indicates 0.5 expected changes per site per branch
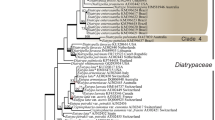
For the DSE-88 group, the analyses of the combined dataset consisting of 93 taxa and 5403 characters showed that the group supported an independent monophyletic clade in Trematosphaeriaceae (Massarineae) as a basal lineage (Fig. 4). The families Trematosphaeriaceae and Morosphaeriaceae together formed a monophyletic group, and the DSE-88 clade was a sister group (PP = 1, ML BS = 83) with species from the genera Halomassarina, Falciformispora, and Trematosphaeria, as well as with the species from the three known genera of Trematosphaeriaceae (Fig. 4). The results of molecular phylogenetic analyses indicated that the DSE-88 isolates collected in the semiarid grasslands represented a novel monospecific genus.
Phylogenetic tree of Fuscosphaeria hungarica isolates and representative species of families in the suborder Massarineae (Pleosporales). Strains presented in this study are shown in bold. The 50% majority rule consensus phylogram inferred from the Bayesian analysis of the combined dataset of four loci (ITS, LSU, SSU, TEF). Bayesian posterior probabilities (≥ 90) are shown before slashes, ML bootstrap support values (≥ 70) are shown after slashes. Leptosphaeria doliolum var. doliolum (CBS 505.75) served as outgroup. Highlighted sections indicate affiliations to families and asterisks indicate unknown clades sensu Tanaka et al. (2015). The scale bar indicates 0.5 expected changes per site per branch
Taxonomy
Fuscosphaeria hungarica D.G. Knapp & Pintye, gen. et sp. nov. (Figs. 2, 4)
MycoBank: MB 835597, MB 835598
Typification: HUNGARY. KISKUNSÁG: semiarid sandy open grassland near Fülöpháza, N46° 52′ E19° 25′, in root of Festuca vaginata, 22 Apr 2014, D.G. Knapp DSE883 (CBS 147250) (holotype 111139BP). GenBank: ITS = MW209054; LSU = MW209059; SSU = MW209065; TEF = MW238843; RPB1 = MW238822; TUB = MW238825.
Etymology: Fusco (from the Latin word fuscus, meaning dark, swarthy) + sphaeria (from the Greek word sphairion, diminutive of sphaira, meaning ball, sphere, referring to its pigmented ascomata-like structures). The species was named “hungarica” because its occurrence solely in one semiarid Hungarian grassland.
Fuscosphaeria hungarica differs from one of its closest phylogenetic neighbors Trematosphaeria pertusa CBS 122368, the ex-type strain of the type species of Trematosphaeria (Treamatosphaeriaceae), by its unique fixed alleles in the LSU, SSU, and TEF loci, which was found based on the alignments of separate loci deposited in TreeBASE as study S26261: LSU positions: 54 (T), 61 (C), 62 (C), 63 (G), 64 (T), 67–70 (deletion), 172 (A), 181 (T), 197 (T), 204 (C), 216 (A), 235 (T), 237 (T), 337 (G), 383 (T), 386 (C), 387 (T), 389 (C), 391 (T), 392 (A), 401 (T), 408 (deletion), 412 (T), 414–415 (deletion), 429 (C), 430 (G), 446 (T), 449 (G), 468 (C), 474 (G), 476 (C), 479 (deletion), 481 (T), 484 (T), 486 (G), 497 (A), 537 (C), 545 (G), 547 (T), 553 (C), 561 (deletion), 564 (T), 571 (A), 578 (C), 582 (C), 663 (A), 669 (A), 674 (A), 690 (G), 708 (T), 709 (C), 710–720 (deletion), 724 (T); SSU positions: 116 (C), 120 (A), 128 (G), 133 (G), 159 (G), 160 (C), 160 (C), 162 (C), 164 (T), 165 (T), 166 (C), 167 (G), 168 (G), 169 (G), 187 (G), 198 (G), 206 (T), 263 (A), 273 (T), 293 (G), 296 (G), 313 (G), 317 (C), 324 (G), 335 (A), 343 (C), 378 (C), 383 (C), 395 (G), 398 (G), 506 (G), 507 (A), 508 (C), 509 (A), 510 (T), 511 (T), 512 (C), 513 (A), 515 (C), 517 (C), 523 (A), 524 (T), 539 (A), 540 (G), 545 (T), 553 (C), 554 (C), 555 (T), 556 (C), 557 (G), 558 (C), 560 (A), 562 (C), 563 (G), 564 (C), 566 (G), 568 (G), 572 (T), 573 (T), 575 (C), 583 (A), 597 (C), 598 (T), 601 (G), 623 (A), 641 (G), 642 (A), 653 (A), 654 (T), 655 (C), 656 (G), 660 (A), 662 (G), 663 (A), 664 (T), 673 (A), 675 (T), 689 (G), 691 (C), 692 (A), 693 (C), 705 (T), 711 (A), 728 (G), 729 (T), 740 (C), 748 (C), 779 (A), 780 (T), 793 (C), 801 (A), 817 (C), 831 (G), 834 (C), 839 (G), 841 (deletion), 912 (deletion), 918 (deletion), 920 (T), 923 (T), 924 (T), 939 (G), 941 (G), 990 (G), 991 (G), 992 (A), 993 (G), 994 (T), 995 (C), 996 (G), 997 (C), 998 (G), 999 (C), 1000 (T), 1001 (T), 1002 (C), 1003 (G), 1004 (C), 1005 (A), 1009 (G), 1012 (C), 1023 (G), 1024 (A), 1025 (A), 1034 (T), 1035 (T), 1037 (T), 1038 (C), 1040 (C), 1043 (G), 1044–1045 (deletion), 1051 (C), 1052 (C), 1055 (A), 1056 (A), 1057 (A), 1059 (A), 1196 (T), 1197 (C), 1222 (deletion), 1223 (G), 1316 (G), 1326 (T), 1329 (G), 1332 (C), 1333 (T), 1388 (A), 1424 (T), 1425 (T), 1454 (T); TEF positions: 24 (C), 30 (G), 37 (C), 55 (T), 91 (C), 94 (C), 115 (G), 116 (A), 118 (G), 127 (C), 133 (T), 152 (A), 169 (G), 172 (C), 174 (T), 176 (C), 178 (C), 184 (C), 190 (A), 196 (G), 208 (T), 217 (T), 220 (A), 226 (C), 241 (A), 250 (C), 256 (C), 263 (A), 283 (G), 284 (C), 289 (G), 290 (A), 331 (G), 334 (C), 338 (T), 339 (C), 341 (T), 343 (T), 352 (T), 355 (G), 358 (C), 361 (A), 364 (C), 367 (T), 372 (G), 386 (A), 394 (C), 397 (T), 409 (C), 415 (A), 418 (C), 421 (T), 439 (C), 448 (C), 454 (G), 483 (C), 491 (T), 493 (C), 511 (C), 517 (T), 538 (A), 544 (C), 562 (G), 569 (A), 570 (C), 571 (T), 572 (G), 578 (C), 580 (C), 589 (T), 607 (T), 619 (C), 634 (T), 637 (C), 643 (T), 649 (T), 652 (T), 655 (T), 673 (T), 685 (C), 688 (A), 712 (T), 715 (C), 733 (G), 745 (T), 769 (T), 778 (T), 781 (T), 793 (T), 802 (G), 832 (C), 835 (G), 865 (C), 868 (T), 895 (C), 898 (A), 916 (C), 922 (C).
Culture characteristics: Colonies on MEA are yellowish grey and flat with sparse aerial mycelium. Strains stain the agar dark brown. On PDA, colonies are dark grey and flat with a pale brown marginal zone, and they stain the agar light brown. Chlamydospores were formed.
Additional material examined: HUNGARY. KISKUNSÁG: semiarid sandy open grassland near Fülöpháza, N46° 52′ E19° 25′, on the roots of Festuca vaginata, 22 Apr 2014, D.G. Knapp DSE881; ibid., DSE882; ibid., DSE884; ibid., DSE885.
Notes: Sterile, depressed, globose, sporocarp-like structures were formed on the surface of autoclaved stinging nettle and submerged in WA media supplemented with minced vegetables or pine needles kept at 20 °C 3 months after inoculation. The change in the color of the WA media with minced vegetables from orange to dark brown was observed.
Ecology and distribution: Sandy grasslands of the Kiskunság region of the Great Hungarian Plain, in the areas dominated by Festuca vaginata and Stipa borysthenica as dominant grasses. Isolates belonging to the genus Fuscosphaeria are root-colonizing fungi associated with F. vaginata in Fülöpháza, Hungary. There is neither an isolate/morph-based nor an uncultured sequence-based report of this species from any other geographical region.
Discussion
In the present study, the DSE-87 isolates representing Delitschia chaetomioides (Delitschiaceae) were first reported as root-colonizing fungi, and a novel species Fuscosphaeria hungarica belonging to a monotypic genus was introduced based on phylogenetic analyses and morphological characters. The species was nested as a basal lineage in the family Trematosphaeriaceae. Although in vitro resynthesis experiments and microscopical investigation of root colonization were not carried out to prove the true endophytic nature of fungi (Grunewaldt-Stöcker and von Alten 2016), in the present study, we considered them to be root-colonizing fungi as they were isolated from healthy tissues of the grass F. vaginata, indicating that they might be considered as fungal root endophytes.
The monogeneric family Delitschiaceae comprises mainly saprobic species inhabiting herbivore dung, wood, or fruits (Kruys et al. 2006; Doveri 2011; Hyde et al. 2013; Jayasiri et al. 2019). Based mainly on the LSU and SSU sequences, Delitschiaceae forms a monophyletic basal lineage of Pleosporales (Hyde et al. 2013; Jayasiri et al. 2019; Haridas et al. 2020). ITS sequencing may be complicated in all species belonging to this family, which was indicated by the low number of sequences deposited online; however, because of the great length and potential variability of this region, ITS sequencing could provide crucial information for species delimitation in this group. The ITS region of these species could also provide misleading information for phylogenetic reconstructions, because the nrDNA sequences can be a primary source of artifacts (Kolařík and Vohník 2018). Nevertheless, obtaining the ITS sequences in combination with the sequences from other loci (e.g., TEF and RPB2) for more Delitschia species could be used to better characterize their lineages and may lead to the separation of the clade comprising the DSE-87 isolates from D. chaetomioides.
Delitschiaceae species are described by sexual morphs, and asexual characters cannot be found in their descriptions. Our DSE-87 isolates belonged to the species Delitschia chaetomioides, which produces embedded, scattered, or clustered perithecia. These ascomata can be black, opaque, globose, or pyriform, and are surrounded by dark hairs. These hairs are brown, septate, flexuous, branched or unbranched with blunt apices, and have thickened walls, up to 1000 μm in length. The asci of D. chaetomioides are eight-spored and cylindrical, and their ascospores are uniseriate, oblong-ellipsoid, at maturity almost black and opaque, each surrounded by a gelatinous layer (Luck-Allen and Cain 1975). In the present study, we also observed dark haired sporocarp-like structures after which the species was named (referring to the typical structures of Chaetomium species). The observed structures were sterile, without asexual or sexual spores, and were almost five times smaller than the usual mature ascomata of D. chaetomioides containing ascospores; however, the morphological characteristics of these morphs as well as the globose shape and abundant appendages or hairs are in accordance with the original description of this species (Luck-Allen and Cain 1975).
D. chaetomioides has been generally reported as a saprobic fungus living on herbivore dung, but it has also been isolated from the shoots of healthy plants, in which it was considered as an endophyte. Khoyratty et al. (2015) isolated D. chaetomioides from the ovaries of Vanilla planifolia and identified it by the LSU sequence. Based on the BLASTN search, one of the closest ITS hits to our D. chaetomioides isolates was an endophyte isolated from the leaves and stems of Trifolium subterraneum from semiarid dehesa ecosystems (grasslands with scattered trees and a herbaceous understory) in Spain (Lledó et al. 2016). The isolates were considered as unidentified fungi, and their sequence was deposited as Dothideomycete sp1 (GenBank accession number KP698360). The same sequences were obtained from the endophytes of another legume species (Ornithopus compressus) during the investigation of the same dehesa sites (Santamaria et al. 2018). Doust et al. (2017) obtained more ITS sequences of D. chaetomioides (GenBank accession number KX611078) from the endophytes of the persian oak (Quercus brantii) originating from a xerophytic, cold-resistant deciduous oak forest. Based on the collection area of our D. chaetomioides isolates and similar sequences from GenBank, we hypothesized that this species can be more strongly associated with the plants from semiarid ecosystems than previously assumed.
Trematosphaeriaceae species are usually saprobes or pathogens (Suetrong et al. 2011; Ahmed et al. 2014). Recently, a couple of taxa were excluded, and based on the LSU, SSU, TEF, and RPB2 regions it was found that this family comprises three genera: Falciformispora, Halomassarina, and Trematosphaeria (Suetrong et al. 2011; Tanaka et al. 2015). Their ascomata are solitary, scattered or produced in groups, initially immersed, becoming erumpent, or semiimmersed, subglobose, black, and have an apex with a short papilla. Their asci are eight-spored, bitunicate, fissitunicate, cylindro-clavate, pedicellate, and possess an ocular chamber. Their ascospores are biseriate to uniseriate, fusiform, hyaline or dark brown, trans-septate, and variously ornamented (Suetrong et al. 2011; Hyde et al. 2013). Only hyphopodia-like structures were reported as anamorphs produced after 6 months in cultures (Suetrong et al. 2011). In the present study, the colonies grown on different media and plant materials were checked periodically for 9 months; no sexual morphs were observed, but only immature or degenerated sporocarp-like structures were found (Fig. 2). Although Fuscosphaeria might represent an incertae sedis clade in a sister position with Trematosphaeriaceae, we considered F. hungarica to be a representative of the most basal group of the family due to its close phylogenetic position and similar morphological characteristics of its ascomata-like structures to the ascomata described in other species from the same family (Suetrong et al. 2011).
Based on a BLAST search of ITS sequences, even if the best hit was below 90% of similarity, the closest hits of F. hungarica were designated as endophytic fungi from different semiarid and arid sites according to GenBank entries and previous publications. During a survey on the effects of grazing on root-associated fungi in a semiarid grassland, Chen et al. (2018) obtained sequences from unidentified fungi (GenBank accession number KX823409) similar to the ITS sequence of F. hungarica. Khidir et al. (2010) detected uncultured root-associated fungi with similar ITS sequences (GenBank accession number FJ362211) from the roots of dominant grasses in semiarid grasslands. Another similar ITS sequence was that of Trematosphaeria sp. (GenBank accession number KY114922) associated with the stem and roots of halophytic species colonizing desert ecosystems (Li et al. 2020). One of the closest hits was another fungal endophyte (GenBank accession number KF887091) originating from Ferula sinkiangensis (Sun et al. 2014).
Root-colonizing endophytic fungi are considered to be asexual (Jumpponen and Trappe 1998); however, sterile, immature, and relatively small ascomata-like structures can rarely be observed under experimental conditions. Currah et al. (1993) observed sterile ascomata-like structures of a DSE species, but without ascospores. Recently, in the case of the worldwide distributed root endophytic pleosporalean genus Darksidea, Knapp et al. (2015) reported induced ascomata production, and some of these structures contained asci and ascospores. Similar to the taxonomical description presented in the present study, novel root endophytic species were generally described without sexual morphs (e.g., Knapp et al. 2015; Ashrafi et al. 2018; Crous et al. 2019; Vohník et al. 2019).
We are not aware of any previously published isolates related to Fuscosphaeria, and D. chaetomioides has also been rarely isolated in previous studies (Khoyratty et al. 2015; Lledó et al. 2016; Santamaria et al. 2018). The reports on the presence of these species are probably biased because of faster growing fungi, which are capable of growing rapidly on media and suppressing other fungal species (Hyde and Soytong 2008). Although based on the results of isolation techniques, these two species are not among the dominant lineages of root-associated fungi (e.g., Knapp et al. 2012, 2015), sequences of both species were found in our databases of the ITS-based metabarcoding study of the soil fungal community in different sites of the grassland near Fülöpháza (Vajna, Knapp, and Kovács, unpublished results), strengthening the idea of their common presence in this semiarid area. Further investigations of non-pathogenic root colonizers of gramineous plants may reveal their broader occurrence on semiarid grasslands, which was found for several root endophytes (Knapp et al. 2019).
References
Ahmed SA, Van De Sande W, Stevens DA, Fahal A, Van Diepeningen A, Menken S, de Hoog G (2014) Revision of agents of black-grain eumycetoma in the order Pleosporales. Persoonia 33:141
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ashrafi S, Knapp DG, Blaudez D, Chalot M, Maciá-Vicente JG, Zagyva I, Dababat AA, Maier W, Kovács GM (2018) Inhabiting plant roots, nematodes, and truffles—Polyphilus, a new helotialean genus with two globally distributed species. Mycologia 110:286–299
Borchsenius F (2009) FastGap 1.2. Software distributed by the authors. [Access date: 04 May 2020]. Available from: http://www.aubot.dk/FastGap_home.htm
Chen T, Zhang Y, Christensen M, Li C, Hou F, Nan Z (2018) Grazing intensity affects communities of culturable root-associated fungi in a semiarid grassland of Northwest China. Land Degrad Dev 29:361–373
Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G (2004) MycoBank: an online initiative to launch mycology into the 21st century. Stud Mycol 50:19–22
Crous PW, Carnegie AJ, Wingfield MJ, Sharma R, Mughini G, Noordeloos ME, Santini A, Shouche YS, Bezerra JD, Dima B et al (2019) Fungal planet description sheets: 868–950. Persoonia 42:291
Currah R, Tsuneda A, Murakami S (1993) Morphology and ecology of Phialocephala fortinii in roots of Rhododendron brachycarpum. Can J Bot 71:1639–1644
Doust NH, Akbarinia M, Safaie N, Yousefzadeh H, Bálint M (2017) Community analysis of Persian oak fungal microbiome under dust storm conditions. Fungal Ecol 29:1–9
Doveri F (2011) Additions to “Fungi Fimicoli Italici”: an update on the occurrence of coprophilous Basidiomycetes and Ascomycetes in Italy with new records and descriptions. Mycosphere 2:331–427
Endresz G, Somodi I, Kalapos T (2013) Arbuscular mycorrhizal colonisation of roots of grass species differing in invasiveness. Community Ecol 14:67–76
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Glass NL, Donaldson GC (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330
Grunewaldt-Stöcker G, von Alten H (2016) Is the root-colonizing endophyte Acremonium strictum an ericoid mycorrhizal fungus? Mycorrhiza 26:429–440
Haridas S, Albert R, Binder M, Bloem J, LaButti K, Salamov A, Andreopoulos B, Baker S, Barry K, Bills G (2020) 101 Dothideomycetes genomes: a test case for predicting lifestyles and emergence of pathogens. Stud Mycol 96:141–153
Hyde KD, Soytong K (2008) The fungal endophyte dilemma. Fungal Divers 33:163–173
Hyde KD, Steinke TS (1996) Two new species of Delitschia from submerged wood. Mycoscience 37:99–102
Hyde KD, Jones EG, Liu J-K, Ariyawansa H, Boehm E, Boonmee S, Braun U, Chomnunti P, Crous PW, Dai D-Q (2013) Families of Dothideomycetes. Fungal Divers 63:1–313
Jayasiri S, Hyde K, Jones E, McKenzie E, Jeewon R, Phillips A, Bhat D, Wanasinghe D, Liu J, Lu Y (2019) Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits
Jumpponen A, Trappe JM (1998) Dark septate endophytes: a review of facultative biotrophic root-colonizing fungi. New Phytol 140:295–310
Jumpponen A, Herrera J, Porras-Alfaro Rudgers J (2017) Biogeography of root-associated fungal endophytes. In: Tedersoo L (eds) Biogeography of Mycorrhizal Symbiosis. Ecological Studies (Analysis and Synthesis), vol 230. Springer, Cham. https://doi.org/10.1007/978-3-319-56363-3_10
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Khidir H, Eudy D, Porras-Alfaro A, Herrera J, Natvig D, Sinsabaugh R (2010) A general suite of fungal endophytes dominate the roots of two dominant grasses in a semiarid grassland. J Arid Environ 74:35–42
Khoyratty S, Dupont J, Lacoste S, Palama TL, Choi YH, Kim HK, Payet B, Grisoni M, Fouillaud M, Verpoorte R (2015) Fungal endophytes of Vanilla planifolia across Réunion Island: isolation, distribution and biotransformation. BMC Plant Biol 15:142
Knapp DG, Pintye A, Kovács GM (2012) The dark side is not fastidious–dark septate endophytic fungi of native and invasive plants of semiarid sandy areas. PLoS One 7(2): e32570
Knapp DG, Kovács GM, Zajta E, Groenewald J, Crous PW (2015) Dark septate endophytic pleosporalean genera from semiarid areas. Persoonia 35:87
Knapp DG, Németh JB, Barry K, Hainaut M, Henrissat B, Johnson J, Kuo A, Lim JHP, Lipzen A, Nolan M (2018) Comparative genomics provides insights into the lifestyle and reveals functional heterogeneity of dark septate endophytic fungi. Sci Rep 8:1–13
Knapp DG, Imrefi I, Boldpurev E, Csíkos S, Akhmetova G, Nagy-Berek PJ, Otgonsuren B, Kovács GM (2019) Root colonizing endophytic fungi of the dominant grass Stipa krylovii from a Mongolian steppe grassland. Front Microbiol 10:2565
Kolařík M, Vohník M (2018) When the ribosomal DNA does not tell the truth: the case of the taxonomic position of Kurtia argillacea, an ericoid mycorrhizal fungus residing among Hymenochaetales. Fungal Biol 122(1):1–18
Kovács GM, Szigetvári C. (2002) Mycorrhizae and other root-associated fungal structures of the plants of a sandy grassland on the Great Hungarian Plain. Phyton 42:211–223
Kruys Å, Eriksson OE, Wedin M (2006) Phylogenetic relationships of coprophilous Pleosporales (Dothideomycetes, Ascomycota), and the classification of some bitunicate taxa of unknown position. Mycol Res 110:527–536
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Li X, Wang J, Zhang S, Wang H, Li X, Li X, Zhang H (2018) Distribution of fungal endophytes in roots of Stipa krylovii across six vegetation types in grassland of northern China. Fungal Ecol 31:47–53
Li J-L, Sun X, Zheng Y, Lü P-P, Wang Y-L, Guo L-D (2020) Diversity and community of culturable endophytic fungi from stems and roots of desert halophytes in Northwest China. MycoKeys 62:75
Liu YJ, Whelen S, Hall BD (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol 16:1799–1808
Lledó S, Santamaría O, Rodrigo S, Poblaciones M (2016) Endophytic mycobiota associated with Trifolium subterraneum growing under semiarid conditions. Ann Appl Biol 168:243–254
Luck-Allen ER, Cain RF (1975) Additions to the genus Delitschia. Can J Bot 53:1827–1887
Lukešová T, Kohout P, Větrovský T, Vohník M (2015) The potential of dark septate endophytes to form root symbioses with ectomycorrhizal and ericoid mycorrhizal middle european forest plants. PLoS One 10(4):e0124752
Mandyam K, Jumpponen A (2005) Seeking the elusive function of the root-colonising dark septate endophytic fungi. Stud Mycol 53:173–189
Mandyam K, Loughin T, Jumpponen A (2010) Isolation and morphological and metabolic characterization of common endophytes in annually burned tallgrass prairie. Mycologia 102:813–821
Matheny PB, Liu YJ, Ammirati JF, Hall BD (2002) Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am J Bot 89:688–698
Nagy LG, Kocsube S, Csanádi Z, Kovacs GM, Petkovits T, Vágvölgyi C, Papp T (2012) Re-mind the gap! Insertion–deletion data reveal neglected phylogenetic potential of the nuclear ribosomal internal transcribed spacer (ITS) of fungi. PLoS One 7:e49794
Porras-Alfaro A, Bayman P (2011) Hidden fungi, emergent properties: endophytes and microbiomes. Annu Rev Phytopathol 49:291–315
Porras-Alfaro A, Herrera J, Sinsabaugh RL, Odenbach KJ, Lowrey T, Natvig DO (2008) Novel root fungal consortium associated with a dominant desert grass. Appl Environ Microbiol 74:2805–2813
Rayner R (1970) A mycological colour chart. CMI and British Mycological Society; Kew, Surrey, England
Rehner SA, Buckley E (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97:84–98
Rehner SA, Samuels GJ (1994) Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res 98:625–634
Rivera-Chávez J, El-Elimat T, Gallagher JM, Graf TN, Fournier J, Panigrahi GK, Deep G, Bunch RL, Raja HA, Oberlies NH (2019) Delitpyrones: α-Pyrone derivatives from a freshwater Delitschia sp. Planta Med 85:62–71
Rodriguez RJ, White JF Jr, Arnold AE, Redman RS (2009) Fungal endophytes: diversity and functional roles. New Phytol 182:314–330
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Santamaria O, Rodrigo S, Lledó S, Poblaciones MJ (2018) Fungal endophytes associated with Ornithopus compressus growing under semiarid conditions. Plant Ecol Divers 11:581–595
Schlegel M, Munsterkotter M, Guldener U, Bruggmann R, Duo A, Hainaut M, Henrissat B, Sieber CMK, Hoffmeister D, Grünig CR (2016) Globally distributed root endophyte Phialocephala subalpina links pathogenic and saprophytic lifestyles. BMC Genomics 17(1):1015
Sieber TN, Grünig CR (2013) Fungal root endophytes. In: Plant Roots: the Hidden Half. Marcel Dekker, New York
Silvestro D, Michalak I (2012) raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 12:335–337
Simmons MP, Ochoterena H, Carr TG (2001) Incorporation, relative homoplasy, and effect of gap characters in sequence-based phylogenetic analyses. Syst Biol 50:454–462
Staden R, Beal KF, Bonfield JK (2000) The Staden package, 1998
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Stiller JW, Hall BD (1997) The origin of red algae: implications for plastid evolution. PNAS 94:4520–4525
Suetrong S, Hyde KD, Zhang Y, Bahkali AH, Jones EG (2011) Trematosphaeriaceae fam. nov. (Dothideomycetes, Ascomycota). Cryptogam Mycol 32:343–358
Sun L, Zhu J, Li X, Shi S, Guo S (2014) Diversity of endophytic fungi associated with Ferula sinkiangensis K. M. Shen. Acta Microbiol Sin 54:936–942
Szabó G, Zimmermann Z, Catorci A, Csontos P, Wichmann B, Szentes S, Barczi A, Penksza K (2017) Comparative study on grasslands dominated by Festuca vaginata and F. pseudovaginata in the Carpathian Basin. Tuexenia 37:415–429
Tanaka K, Hirayama K, Yonezawa H, Sato G, Toriyabe A, Kudo H, Hashimoto A, Matsumura M, Harada Y, Kurihara Y (2015) Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud Mycol 82:75–136
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246
Vohník M, Borovec O, Kolaříková Z, Sudová R, Réblová M (2019) Extensive sampling and high-throughput sequencing reveal Posidoniomycesatricolor gen. et sp. nov. (Aigialaceae, Pleosporales) as the dominant root mycobiont of the dominant Mediterranean seagrass Posidoniaoceanica. MycoKeys 55:59
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocol Guide Methods Appl 18:315–322
Wilson D (1995) Endophyte—the evolution of a term, and clarification of its use and definition. Oikos 73:274–276
Young ND, Healy J (2003) GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinform 4:6
Yuan Z, Druzhinina IS, Wang X, Zhang X, Peng L, Labbé J (2019). Insight into a highly polymorphic endophyte isolated from the roots of the halophytic seepweed Suaeda salsa: Laburnicola rhizohalophila sp. nov. (Didymosphaeriaceae, Pleosporales). Fungal Biol 124:327–337
Zhang Y, Crous PW, Schoch CL, Hyde KD (2012) Pleosporales. Fungal Divers 53:1–221
Acknowledgments
We thank Gábor M. Kovács for the discussions and comments on the manuscript.
Funding
Open access funding provided by Eötvös Loránd University. This research was supported by the National Research, Development and Innovation Office, Hungary (NKFIH KH-130401), the ELTE Institutional Excellence Program supported by the National Research, Development and Innovation Office (NKFIH-1157-8/2019-DT), and the János Bolyai Research Scholarship of the Hungarian Academy of Sciences to Dániel G. Knapp.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. All the data and materials used in the publication are deposited in public databases and culture collections. All authors contributed to the study conception and design. Material preparation, data collection, and analyses were performed and the manuscript was written by Alexandra Pintye and Dániel G. Knapp. All authors commented on previous versions of the manuscript, then read and approved the final manuscript.
Additional information
Section Editor: Gerhard Rambold
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pintye, A., Knapp, D.G. Two pleosporalean root-colonizing fungi, Fuscosphaeria hungarica gen. et sp. nov. and Delitschia chaetomioides, from a semiarid grassland in Hungary. Mycol Progress 20, 39–50 (2021). https://doi.org/10.1007/s11557-020-01655-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-020-01655-8