Abstract
Purpose
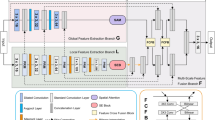
Deep learning has recently shown its outstanding performance in biomedical image semantic segmentation. Most biomedical semantic segmentation frameworks comprise the encoder–decoder architecture directly fusing features of the encoder and the decoder by the way of skip connections. However, the simple fusion operation may neglect the semantic gaps which lie between these features in the decoder and the encoder, hindering the effectiveness of the network.
Methods
Dense gate network (DG-Net) is proposed for biomedical image segmentation. In this model, the Gate Aggregate structure is utilized to reduce the semantic gaps between features in the encoder and the corresponding features in the decoder, and the gate unit is used to reduce the categorical ambiguity as well as to guide the low-level high-resolution features to recover semantic information. Through this method, the features could reach a similar semantic level before fusion, which is helpful for reducing semantic gaps, thereby producing accurate results.
Results
Four medical semantic segmentation experiments, based on CT and microscopy images datasets, were performed to evaluate our model. In the cross-validation experiments, the proposed method achieves IOU scores of 97.953%, 89.569%, 81.870% and 76.486% on these four datasets. Compared with U-Net and MultiResUNet methods, DG-Net yields a higher average score on IOU and Acc.
Conclusion
The DG-Net is competitive with the baseline methods. The experiment results indicate that Gate Aggregate structure and gate unit could improve the performance of the network by aggregating features from different layers and reducing the semantic gaps of features in the encoder and the decoder. This has potential in biomedical image segmentation.







Similar content being viewed by others
References
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: CVPR. arxiv:1411.4038
Zhao H, Shi J, Qi X, Wang X, Jia J (2017) Pyramid scene parsing network. In: CVPR. arxiv:1411.4038
Chen L-C, Zhu Y, Papandreou G, Schroff F, Adam H (2018) Encoder–decoder with atrous separable convolution for semantic image segmentation. In: CVPR. arxiv:1802.0ds2611fds
Yu C, Wang J, Peng C, Gao C, Yu G, Sang N (2018) BiSeNet: bilateral segmentation network for real-time semantic segmentation. In: ECCV. arxiv:1808.00897
Chaurasia A, Culurciello E (2017) LinkNet: exploiting encoder representations for efficient semantic segmentation. In: VCIP 2017. arxiv:1707.0378
Pohlen T, Hermans A, Mathias M, Leibe B (2017) Full-resolution residual networks for semantic segmentation in street scenes. In: CVPR. arxiv:1611.08323
Meletis Panagiotis, Dubbelman Gijs (2018) Training of convolutional networks on multiple heterogeneous datasets for street scene semantic segmentation. IEEE Intell Veh Symp (IV) 2018:1045–1105
Fu H, Xu Y, Wing D, Wong K, Liu J (2016) Retinal vessel segmentation via deep learning network and fully-connected conditional random fields. In: 2016 IEEE 13th international symposium on biomedical imaging (ISBI). IEEE
Dou Q, Chen H, Jin Y, Yu L, Qin J, Heng P-A (2016) 3D Deeply supervised network for automatic liver segmentation from CT volumes. In: CVPR 2016. arxiv:1607.00582
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. arxiv:1505.04597
Beers A, Chang K, Brown J, Sartor E, Mammen CP, Gerstner E, Rosen B, Kalpathy-Cramer J (2017) Sequential 3D U-Nets for biologically-informed brain tumor segmentation. In: CVPR 2017. arxiv:1709.02967
Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B, Glocker B, Rueckert D (2018). Attention U-Net: learning where to look for the pancreas. In: 1st conference on medical imaging with deep learning (MIDL 2018). arxiv:1804.03999
Kamrul Hasan SM, Linte CA (2019) U-NetPlus: a modified encoder–decoder U-Net architecture for semantic and instance segmentation of surgical instrument. arixv:1902.08994
Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C (2016) The importance of skip connections in biomedical image segmentation. arxiv:1608.04117
Zhang Z, Zhang X, Peng C, Cheng D, Sun J (2018) ExFuse: enhancing feature fusion for semantic segmentation. In: European conference on computer vision. Springer, Cham. arxiv:1804.03821
Ibtehaz N, Sohel Rahman M (2019) MultiResUNet: rethinking the U-Net architecture for multimodal biomedical image segmentation. arxiv:1902.04049
Yu F, Wang D, Shelhamer E, Darrell T (2018) Deep layer aggregation. In: CVPR 2018. arxiv:1707.06484
Huang G, Liu Z, van der Maaten L (2016) Densely connected convolutional networks. 2016. In: CVPR. arxiv:1608.06993
Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J (2018) UNet++: a nested U-Net architecture for medical image segmentation. In: LMIA. arxiv:1807.10165
Islam MA, Rochan M, Bruce NDB, Wang Y (2017) Gated feedback refinement network for dense image labeling. In: CVPR
Ulman V, Maška M, Magnusson KEG, Ronneberger O, Haubold C, Harder N, Matula P, Matula P, Svoboda D, Radojevic M, Smal I, Rohr K, Jaldén J, Blau HM, Dzyubachyk O, Lelieveldt B, Xiao P, Li Y, Cho S-Y, Dufour AC, Olivo-Marin J-C, Reyes-Aldasoro CC, Solis-Lemus JA et al (2017) An objective comparison of cell-tracking algorithms. Nat Methods 14:1141–1152
Maška M, Ulman V, Svoboda D, Matula P, Matula P, Ederra C, Urbiola A, Españ T, Venkatesan S, Balak DMW, Karas P, Bolcková T, Štreitová M, Carthel C, Coraluppi S, Harder N, Rohr K, Magnusson KEG, Jaldén J, Blau HM, Dzyubachyk O, Křížek P, Hagen GM, Escuredo DP, Jimenez-Carretero D, Ledesma-Carbayo MJ, Muñoz-Barrutia A, Meijering E, Kozubek M, Ortiz-de-Solorzano C (2014) A benchmark for comparison of cell tracking algorithms. Bioinformatics 30(11):1609–1617
Bloice Marcus D, Roth Peter M, Holzinger A (2019) Biomedical image augmentation using Augmentor. Bioinformatics 35(21):4522–4524. https://doi.org/10.1093/bioinformatics/btz259
Acknowledgements
The authors are grateful to Dr. Jerod Michel for his help in polishing the language.
Funding
This study was funded by the National Natural Science Foundation of China (Grant Nos. 61773205, 61171059).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The mouse CT images were scanned in Southeast University, and the experiment was approved by the Animal Ethics Committee of Southeast University. All applicable Animal Ethics Committee of Southeast University guidelines for the care and use of animals were followed.
Informed consent
This article does not contain patient data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, D., Chen, C., Li, J. et al. Dense gate network for biomedical image segmentation. Int J CARS 15, 1247–1255 (2020). https://doi.org/10.1007/s11548-020-02138-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-020-02138-7