Abstract
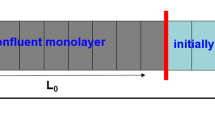
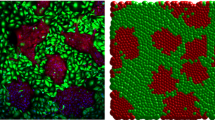
Standard differential equation-based models of collective cell behaviour, such as the logistic growth model, invoke a mean-field assumption which is equivalent to assuming that individuals within the population interact with each other in proportion to the average population density. Implementing such assumptions implies that the dynamics of the system are unaffected by spatial structure, such as the formation of patches or clusters within the population. Recent theoretical developments have introduced a class of models, known as moment dynamics models, which aim to account for the dynamics of individuals, pairs of individuals, triplets of individuals, and so on. Such models enable us to describe the dynamics of populations with clustering, however, little progress has been made with regard to applying moment dynamics models to experimental data. Here, we report new experimental results describing the formation of a monolayer of cells using two different cell types: 3T3 fibroblast cells and MDA MB 231 breast cancer cells. Our analysis indicates that the 3T3 fibroblast cells are relatively motile and we observe that the 3T3 fibroblast monolayer forms without clustering. Alternatively, the MDA MB 231 cells are less motile and we observe that the MDA MB 231 monolayer formation is associated with significant clustering. We calibrate a moment dynamics model and a standard mean-field model to both data sets. Our results indicate that the mean-field and moment dynamics models provide similar descriptions of the 3T3 fibroblast monolayer formation whereas these two models give very different predictions for the MDA MD 231 monolayer formation. These outcomes indicate that standard mean-field models of collective cell behaviour are not always appropriate and that care ought to be exercised when implementing such a model.




Similar content being viewed by others
References
Baker, R. E., & Simpson, M. J. (2010). Correcting mean-field approximations for birth–death–movement processes. Phys. Rev. E, 82, 041905.
Binder, B. J., & Landman, K. A. (2011). Quantifying evenly distributed states in exclusion and nonexclusion processes. Phys. Rev. E, 83, 041914.
Binder, B. J., Hackett-Jones, E. J., Tuke, J., & Landman, K. A. (2011). A modified Pólya urn process and an index for spatial distributions with volume exclusion. ANZIAM J., 53, 122–133.
Binder, B. J., Landman, K. A., Newgreen, D. F., Simkin, J. E., Takahashi, Y., & Zhang, D. (2012). Spatial analysis of multi-species exclusion processes: application to neural crest cell migration in the embryonic gut. Bull. Math. Biol., 74, 474–490.
Bolker, B., & Pacala, S. W. (1997). Using moment equations to understand stochastically-driven spatial pattern formation in ecological systems. Theor. Popul. Biol., 52, 179–197.
Bruna, M., & Chapman, S. J. (2012). Excluded-volume effects in the diffusion of hard spheres. Phys. Rev. E, 85, 011103.
Cai, A. Q., Landman, K. A., & Hughes, B. D. (2007). Multi-scale modeling of a wound-healing cell migration assay. J. Theor. Biol., 245, 576–594.
Cailleau, R., Young, R., Olivé, M., & Reeves WJ, Jr. (1974). Breast tumor cell lines from pleural effusions. J. Natl. Cancer Inst., 53, 661–674.
Codling, E. A., Plank, M. J., & Benhamou, S. (2008). Random walk models in biology. J. R. Soc. Interface, 5, 813–834.
Dangerfield, C. E., Ross, J. V., & Keeling, M. J. (2008). Integrating stochasticity and network structure into an epidemic model. J. R. Soc. Interface, 6, 761–774.
Deroulers, C., Aubert, M., Badoual, M., & Grammaticos, B. (2009). Modeling tumor cell migration: from microscopic to macroscopic models. Phys. Rev. E, 79, 031917.
Dyson, L., Maini, P. K., & Baker, R. E. (2012). Macroscopic limits of individual-based models for motile cell populations with volume exclusion. Phys. Rev. E, 86, 031903.
Gillespie, D. T. (1977). Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem., 81, 2340–2631.
Hackett-Jones, E. J., Landman, K. A., Newgreen, D. F., & Zhang, D. (2011). On the role of differential adhesion in gangliogenesis in the enteric nervous system. J. Theor. Biol., 287, 148–159.
Hackett-Jones, E. J., Davies, K. J., Binder, B. J., & Landman, K. A. (2012). Generalized index for spatial data sets as a measure of complete spatial randomness. Phys. Rev. E, 85, 061908.
Hansen, J.-P., & McDonald, I. R. (2006). Theory of simple liquids (3rd edn.). London: Elsevier/Academic Press.
Hughes, B. D. (1995). Random walks and random environments (Vol. 1). Oxford: Oxford University Press.
Illian, J., Penttinen, A., Stoyan, H., & Stoyan, D. (2008). Statistical analysis and modelling of spatial point patterns. Chichester: Wiley.
Johnston, S. T., Simpson, M. J., & Baker, R. E. (2012). Mean-field descriptions of collective migration with strong adhesion. Phys. Rev. E, 85, 051922.
Keeling, M. J., Rand, D. A., & Morris, A. J. (1997). Correlation models for childhood epidemics. Proc. R. Soc. Lond. B, 264, 1149–1156.
Lah, G. J., & Key, B. (2012). Novel roles of the chemorepellent axon guidance molecule RGMa in cell migration and adhesion. Mol. Cell. Biol., 32, 968–980.
Law, R., & Dieckmann, U. (2000). A dynamical system for neighbourhoods in plant communities. Ecology, 81, 2137–2148.
Law, R., Murrell, D. J., & Dieckmann, U. (2003). Population growth in space and time: spatial logistic equations. Ecology, 84, 252–262.
Law, R., Illian, J., Burslem, D. F. R. P., Gratzer, G., Gunatilleke, C. V. S., & Gunatilleke, A. U. N. (2009). Ecological information from spatial patterns of plants: insights from point process theory. J. Ecol., 97, 616–628.
Liggett, T. M. (1999). Stochastic interacting systems: contact, voter and exclusion processes. Berlin: Springer.
Mai, J., Kuzovkov, V. N., & von Niessen, W. (1993). A theoretical stochastic model for the A+1/2B 2→0 reaction. J. Chem. Phys., 98, 10017–10025.
Mai, J., Kuzovkov, V. N., & von Niessen, W. (1994). A general stochastic model for the description of surface reaction systems. Physica A, 203, 298–315.
Maini, P. K., McElwain, D. L. S., & Leavesley, D. I. (2004a). Traveling wave model to interpret a wound-healing cell migration assay for human peritoneal mesothelial cells. Tissue Eng., 10, 475–482.
Maini, P. K., McElwain, D. L. S., & Leavesley, D. (2004b). Travelling waves in a wound healing assay. Appl. Math. Lett., 17, 575–580.
Massey, S. C., Assanah, M. C., Lopez, K. A., Canoll, P., & Swanson, K. R. (2012). Glial progenitor cell recruitment drives aggressive glioma growth: mathematical and experimental modelling. J. R. Soc. Interface, 9, 1757–1766.
Murray, J. D. (2002). Mathematical biology I: an introduction (3rd edn.). Heidelberg: Springer.
Murrell, D. J., Dieckmann, U., & Law, R. (2004). On moment closures for population dynamics in continuous space. J. Theor. Biol., 229, 421–432.
Penington, C. J., Hughes, B. D., & Landman, K. A. (2011). Building macroscale models from microscale probabilistic models: a general probabilistic approach for nonlinear diffusion and multispecies phenomena. Phys. Rev. E, 84, 041120.
Penington, C. J., Korvasová, K., Hughes, B. D., & Landman, K. A. (2012). Collective motion of dimers. Phys. Rev. E, 86, 051909.
Phelps, J. H., & Tucker, C. L. (2006). Lagrangian particle calculations of distributive mixing: limitations and applications. Chem. Eng. Sci., 61, 6826–6836.
Plank, M. J., & Simpson, M. J. (2012). Models of collective cell behaviour with crowding effects: comparing lattice-based and lattice-free approaches. J. R. Soc. Interface, 9, 2983–2996.
Raghib, M., Hill, N. A., & Dieckmann, U. (2011). A multiscale maximum entropy moment closure for locally regulated space—time point process models of population dynamics. J. Math. Biol., 62, 605–653.
Sengers, B. G., Please, C. P., & Oreffo, R. OC. (2007). Experimental characterization and computational modelling of two-dimensional cell spreading for skeletal regeneration. J. R. Soc. Interface, 4, 1107–1117.
Sharkey, K. J. (2008). Deterministic epidemiological models at the individual level. J. Math. Biol., 57, 311–331.
Sharkey, K. J. (2011). Deterministic epidemic models on contact networks: correlations and unbiological terms. Theor. Popul. Biol., 79, 115–129.
Sherratt, J. A., & Murray, J. D. (1990). Models of epidermal wound healing. Proc. R. Soc. Lond. B, 241, 29–36.
Shiozaki, H., Tahara, H., Oka, H., Miyata, M., Kobayashi, K., Tamura, S., Iihara, K., Doki, Y., Hirano, S., Takeichi, M., & Mori, T. (1991). Expression of immunoreactive E-Cadherin adhesion molecules in human cancers. Am. J. Pathol., 139, 17–23.
Simpson, M. J., & Baker, R. E. (2011). Corrected mean-field models for spatially-dependent advection–diffusion–reaction phenomena. Phys. Rev. E, 83, 051922.
Simpson, M. J., Landman, K. A., Hughes, B. D., & Fernando, A. E. (2010a). A model for mesoscale patterns in motile populations. Physica A, 389, 1412–1424.
Simpson, M. J., Landman, K. A., & Hughes, B. D. (2010b). Cell invasion with proliferation mechanisms motivated by time-lapse data. Physica A, 389, 3779–3790.
Simpson, M. J., Baker, R. E., & McCue, S. W. (2011). Models of collective cell spreading with variable cell aspect ratio: a motivation for degenerate diffusion models. Phys. Rev. E, 83, 021901.
Simpson, M. J., Treloar, K. K., Binder, B. J., Haridas, P., Manton, K., Leavesley, D. I., McElwain, D. L. S., & Baker, R. E. (2013). Quantifying the roles of cell motility and cell proliferation in a circular barrier assay. J. R. Soc. Interface, 10, 20130007.
Singer, A. (2004). Maximum entropy formulation of the Kirkwood superposition approximation. J. Chem. Phys., 121, 3657.
Suzuki, T., Fujikura, K., Higashiyama, T., & Takata, K. (1997). DNA staining for fluorescence and laser confocal microscopy. J. Histochem. Cytochem., 45, 49–53.
Swanson, K. R., Bridge, C., Murray, J. D., & Alvord, E. C. Jr. (2003). Virtual and real brain tumors: using mathematical modeling to quantify glioma growth and invasion. J. Neurol. Sci., 216, 1–10.
Takeichi, M. (1991). Cadherin cell adhesion receptors as a morphogenetic regulator. Science, 251, 1451–1555.
Tamm, I., Kikuchi, Y., Cardinale, I., & Krueger, J. G. (1994). Cell-adhesion-disrupting action of interleukin 6 in human ductal breast carcinoma cells. Proc. Natl. Acad. Sci. USA, 91, 3329–3333.
Todaro, G. J., & Green, H. (1963). Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol., 17, 299–313.
Tremel, A., Cai, A. Q., Tirtaatmadja, N., Hughes, B. D., Stevens, G. W., Landman, K. A., & O’Connor, A. J. (2009). Cell migration and proliferation during monolayer formation and wound healing. Chem. Eng. Sci., 64, 247–253.
Young, W. R., Roberts, A. J., & Stuhne, G. (2001). Reproductive pair correlations and the clustering of organisms. Nature, 412, 328–331.
Acknowledgements
We appreciate the advice and assistance of Associate Professor David Leavesley, Dr. Kerry Manton and Dr Leo de Boer. This research is supported by the Australian Research Council Discovery Project DP120100551, and the 2011 International Exchange Scheme funded by the Royal Society.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Simpson, M.J., Binder, B.J., Haridas, P. et al. Experimental and Modelling Investigation of Monolayer Development with Clustering. Bull Math Biol 75, 871–889 (2013). https://doi.org/10.1007/s11538-013-9839-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-013-9839-0