Abstract
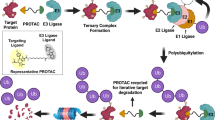
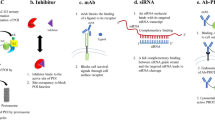
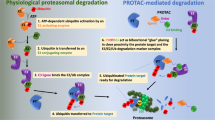
PROteolysis Targeting Chimeras (PROTACs) are a family of heterobifunctional small molecules that specifically target cellular proteins for degradation. Given that their mode of action is distinct from that of small-molecule inhibitors widely used in clinical practice, PROTACs have the potential to improve current cancer therapies. Multiple studies have suggested that PROTACs exhibit enhanced pharmacodynamics and reduced toxicity both in vitro and in vivo compared to clinically relevant small-molecule kinase inhibitors. In addition, PROTACs have been reported to be less prone to mutation-mediated drug resistance in specific disease settings. Since its development in 2001, the field of targeted protein degradation, in which PROTACs are used, has expanded rapidly. However, earlier studies focused on the advancement of the technology itself, while preclinical and clinical data on the disease-modifying effect of PROTACs have only recently been reported. As preclinical and clinical evidence accumulates, the efficacy of PROTACs as targeted therapeutics—distinct from that of small-molecule kinase inhibitors—suggests potential translational benefit in the clinical setting. In this short review, we aim to describe translational potentials of PROTACs. We offer our perspectives as practicing oncologists on the preclinical and clinical data on PROTACs as novel therapeutics for both solid and hematological malignancies.



Similar content being viewed by others
References
Bhullar KS, Lagarón NO, McGowan EM, Parmar I, Jha A, Hubbard BP, et al. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol Cancer. 2018;17(1):48.
Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513.
Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29.
Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18(6):579–86.
Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci USA. 2001;98(15):8554–9.
Cromm PM, Crews CM. The proteasome in modern drug discovery: second life of a highly valuable drug target. ACS Cent Sci. 2017;3:830–8.
Burslem GM, Crews CM. Small-molecule modulation of protein homeostasis. Chem Rev. 2017;117:11269–301.
Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348(6241):1376–81.
Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AMK, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci USA. 2016;113(26):7124–9.
Scheepstra M, Hekking KFW, van Hijfte L, Folmer RHA. Bivalent ligands for protein degradation in drug discovery. Comput Struct Biotechnol J. 2019;17:160–76.
Bai L, Zhou B, Yang C-Y, Ji J, McEachern D, Przybranowski S, et al. Targeted degradation of BET proteins in triple-negative breast cancer. Cancer Res. 2017;77(9):2476–87.
Gechijian LN, Buckley DL, Lawlor MA, Reyes JM, Paulk J, Ott CJ, et al. Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nat Chem Biol. 2018;14(4):405–12.
Bondeson DP, Mares A, Smith IED, Ko E, Campos S, Miah AH, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol. 2015;11(8):611–7.
Zhang X, Crowley VM, Wucherpfennig TG, Dix MM, Cravatt BF. Electrophilic PROTACs that degrade nuclear proteins by engaging DCAF16. Nat Chem Biol. 2019;5(7):737–46.
He Y, Zhang X, Chang J, Kim HN, Zhang P, Wang Y, et al. Using proteolysis-targeting chimera technology to reduce navitoclax platelet toxicity and improve its senolytic activity. Nat Commun. 2020;11(1):1996.
Liao H, Li X, Zhao L, Wang Y, Wang X, Wu Y, et al. A PROTAC peptide induces durable β-catenin degradation and suppresses Wnt-dependent intestinal cancer. Cell Discov. 2020;6:35.
Cromm PM, Samarasinghe KTG, Hines J, Crews CM. Addressing kinase-independent functions of Fak via PROTAC-mediated degradation. J Am Chem Soc. 2018;140(49):17019–26.
Murtuza A, Bulbul A, Shen JP, Keshavarzian P, Woodward BD, Lopez-Diaz FJ, et al. Novel third-generation EGFR tyrosine kinase inhibitors and strategies to overcome therapeutic resistance in lung cancer. Cancer Res. 2019;79(4):689–98.
Cheng M, Yu X, Lu K, Xie L, Wang L, Meng F, et al. Discovery of potent and selective epidermal growth factor receptor (EGFR) bifunctional Small-molecule degraders. J Med Chem. 2020;63(3):1216–32.
Burslem GM, Smith BE, Lai AC, Jaime-Figueroa S, McQuaid DC, Bondeson DP, et al. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem Biol. 2018;25(1):67-77.e3.
Han X-R, Chen L, Wei Y, Yu W, Chen Y, Zhang C, et al. Discovery of selective small molecule degraders of BRAF-V600E. J Med Chem. 2020;63:4069–80.
Posternak G, Tang X, Maisonneuve P, Jin T, Lavoie H, Daou S, et al. Functional characterization of a PROTAC directed against BRAF mutant V600E. Nat Chem Biol. 2020;16(11):1170–8.
O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13(7):417–30.
Brand M, Jiang B, Bauer S, Donovan KA, Liang Y, Wang ES, et al. Homolog-selective degradation as a strategy to probe the function of CDK6 in AML. Cell Chem Biol. 2019;26(2):300-306.e9.
Tovell H, Testa A, Zhou H, Shpiro N, Crafter C, Ciulli A, et al. Design and characterization of SGK3-PROTAC1, an isoform specific SGK3 kinase PROTAC degrader. ACS Chem Biol. 2019;14(9):2024–34.
Hughes SJ, Ciulli A. Molecular recognition of ternary complexes: a new dimension in the structure-guided design of chemical degraders. Essays Biochem. 2017;61(5):505–16.
Gadd MS, Testa A, Lucas X, Chan K-H, Chen W, Lamont DJ, et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol. 2017;13(5):514–21.
Buhimschi AD, Armstrong HA, Toure M, Jaime-Figueroa S, Chen TL, Lehman AM, et al. Targeting the C481S ibrutinib-resistance mutation in Bruton’s tyrosine kinase using PROTAC-mediated degradation. Biochemistry. 2018;57(26):3564–75.
Bondeson DP, Smith BE, Burslem GM, Buhimschi AD, Hines J, Jaime-Figueroa S, et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem Biol. 2018;25(1):78-87.e5.
Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18(6):683–95.
Chen H, Chen F, Pei S, Gou S. Pomalidomide hybrids act as proteolysis targeting chimeras: Synthesis, anticancer activity and B-Raf degradation. Bioorg Chem. 2019;87:191–9.
Nowak RP, Deangelo SL, Buckley D, He Z, Donovan KA, An J, et al. Plasticity in binding confers selectivity in ligand-induced protein degradation article. Nat Chem Biol. 2018;14(7):706–14.
Zorba A, Nguyen C, Xu Y, Starr J, Borzilleri K, Smith J, et al. Delineating the role of cooperativity in the design of potent PROTACs for BTK. Proc Natl Acad Sci USA. 2018;115(31):E7285–92.
Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. Cell Res. 2008;18(4):443–51.
Bai L, Zhou H, Xu R, Zhao Y, Chinnaswamy K, McEachern D, et al. A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell. 2019;36(5):498-511.e17.
Kung JE, Jura N. Structural basis for the non-catalytic functions of protein kinases. Structure. 2016;24(1):7–24.
Potjewyd F, Turner AMW, Beri J, Rectenwald JM, Norris-Drouin JL, Cholensky SH, et al. Degradation of polycomb repressive complex 2 with an EED-targeted bivalent chemical degrader. Cell Chem Biol. 2020;27(1):47-56.e15.
Farnaby W, Koegl M, Roy MJ, Whitworth C, Diers E, Trainor N, et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat Chem Biol. 2019;15:672–80.
Martin LA, Ribas R, Simigdala N, Schuster E, Pancholi S, Tenev T, et al. Discovery of naturally occurring ESR1 mutations in breast cancer cell lines modelling endocrine resistance. Nat Commun. 2017;8(1):1865.
Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–45.
Gonzalez TL, Hancock M, Sun S, Gersch CL, Larios JM, David W, et al. Targeted degradation of activating estrogen receptor α ligand-binding domain mutations in human breast cancer. Breast Cancer Res Treat. 2020;180:611–22.
Hu J, Hu B, Wang M, Xu F, Miao B, Yang CY, et al. Discovery of ERD-308 as a highly potent proteolysis targeting chimera (PROTAC) degrader of estrogen receptor (ER). J Med Chem. 2019;62(3):1420–42.
Stathis A, Bertoni F. BET proteins as targets for anticancer treatment. Cancer Discov. 2018;8(1):24–36.
Lee BY, Timpson P, Horvath LG, Daly RJ. FAK signaling in human cancer as a target for therapeutics. Pharmacol Ther. 2015;146:132–49.
Landesman-Bollag E, Romieu-Mourez R, Song DH, Sonenshein GE, Cardiff RD, Seldin DC. Protein kinase CK2 in mammary gland tumorigenesis. Oncogene. 2001;20(25):3247–57.
Chen H, Chen F, Liu N, Wang X, Gou S. Chemically induced degradation of CK2 by proteolysis targeting chimeras based on a ubiquitin–proteasome pathway. Bioorg Chem. 2018;81:536–44.
Pinna LA. Protein kinase CK2: a challenge to canons. J Cell Sci. 2002;115(Pt 20):3873–8.
Flanagan J, Qian Y, Gough S, Andreoli M, Bookbinder M, Cadelina G, et al. ARV-471, an oral estrogen receptor PROTAC degrader f or breast cancer. Cancer Res. 2019;79(4_suppl):P5-04-18.
Clinical trial of ARV-471 in patients with ER+/HER2- locally advanced or metastatic breast cancer - full text view - ClinicalTrials.gov [Internet]. https://clinicaltrials.gov/ct2/show/NCT04072952#studydesign
Taylor I. Moving PROTAC® protein degraders from the laboratory to the clinic. 2019. https://ir.arvinas.com/static-files/ce10f577-753f-4063-af5e-403f11f75128.
Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–82.
Kregel S, Malik R, Asangani IA, Wilder-Romans K, Rajendiran T, Xiao L, et al. Functional and mechanistic interrogation of BET bromodomain degraders for the treatment of metastatic castration-resistant prostate cancer. Clin Cancer Res. 2019;25(13):4038–48.
Neklesa T, Snyder LB, Willard RR, Vitale N, Pizzano J, Gordon DA, et al. ARV-110: an oral androgen receptor PROTAC degrader for prostate cancer. J Clin Oncol. 2019;37(7_suppl):259.
Petrylak DP, Gao X, Vogelzang NJ, Garfield MH, Taylor I, Dougan Moore M, et al. First-in-human phase I study of ARV-110, an androgen receptor (AR) PROTAC degrader in patients (pts) with metastatic castrate-resistant prostate cancer (mCRPC) following enzalutamide (ENZ) and/or abiraterone (ABI). J Clin Oncol. 2020;38(15_suppl):3500.
Bond MJ, Chu L, Nalawansha DA, Li K, Crews CM. Targeted degradation of oncogenic KRAS G12C by VHL-recruiting PROTACs. ACS Cent Sci. 2020;6(8):1367–75.
Jang J, To C, De Clercq DJH, Park E, Ponthier CM, Shin BH, et al. Mutant-selective allosteric EGFR degraders are effective against a broad range of drug-resistant mutations. Angew Chemie. 2020;59:14481.
Sun N, Ren C, Kong Y, Zhong H, Chen J, Li Y, et al. Development of a brigatinib degrader (SIAIS117) as a potential treatment for ALK positive cancer resistance. Eur J Med Chem. 2020;193:112190.
Zhang C, Han XR, Yang X, Jiang B, Liu J, Xiong Y, et al. Proteolysis targeting chimeras (PROTACs) of anaplastic lymphoma kinase (ALK). Eur J Med Chem. 2018;151:304–14.
Powell CE, Gao Y, Tan L, Donovan KA, Nowak RP, Loehr A, et al. Chemically induced degradation of anaplastic lymphoma kinase (ALK). J Med Chem. 2018;61(9):4249–55.
Kang CH, Lee DH, Lee CO, Du Ha J, Park CH, Hwang JY. Induced protein degradation of anaplastic lymphoma kinase (ALK) by proteolysis targeting chimera (PROTAC). Biochem Biophys Res Commun. 2018;505(2):542–7.
Zhang S, Anjum R, Squillace R, Nadworny S, Zhou T, Keats J, et al. The potent ALK inhibitor brigatinib (AP26113) overcomes mechanisms of resistance to first- and second-generation ALK inhibitors in preclinical models. Clin Cancer Res. 2016;22(22):5527–38.
Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated reactivation of MAPK signaling contributes to insensitivity of BRAF-mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2(3):227–35.
Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7.
Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22(20):2755–66.
Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–8.
Otto C, Schmidt S, Kastner C, Denk S, Kettler J, Müller N, et al. Targeting bromodomain-containing protein 4 (BRD4) inhibits MYC expression in colorectal cancer cells. Neoplasia. 2019;21(11):1110–20.
Cheng J, Li Y, Wang X, Dong G, Sheng C. Discovery of novel PDEδ degraders for the treatment of KRAS mutant colorectal cancer. J Med Chem. 2020;63(14):7892–905.
Burslem GM, Schultz AR, Bondeson DP, Eide CA, Stevens SLS, Druker BJ, et al. Targeting BCR-ABL1 in chronic myeloid leukemia by PROTAC-mediated targeted protein degradation. Cancer Res. 2019;79(18):4744–53.
Zhou H, Xu R. Leukemia stem cells: the root of chronic myeloid leukemia. Protein Cell. 2015;6(6):403–12.
Zhao Q, Ren C, Liu L, Chen J, Shao Y, Sun N, et al. Discovery of SIAIS178 as an effective BCR-ABL degrader by recruiting von Hippel-Lindau (VHL) E3 ubiquitin ligase. J Med Chem. 2019;62(20):9281–98.
Yang Y, Gao H, Sun X, Sun Y, Qiu Y, Weng Q, et al. Global PROTAC toolbox for degrading BCR-ABL overcomes drug-resistant mutants and adverse effects. J Med Chem. 2020;63(15):8567–83.
Vangamudi B, Paul TA, Shah PK, Kost-Alimova M, Nottebaum L, Shi X, et al. The SMARCA2/4 ATPase domain surpasses the bromodomain as a drug target in SWI/SNF-mutant cancers: Insights from cDNA rescue and PFI-3 inhibitor studies. Cancer Res. 2015;75(18):3865–78.
Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27(24):2648–62.
Qin C, Hu Y, Zhou B, Fernandez-Salas E, Yang CY, Liu L, et al. Discovery of QCA570 as an exceptionally potent and efficacious proteolysis targeting chimera (PROTAC) degrader of the bromodomain and extra-terminal (BET) proteins capable of inducing complete and durable tumor regression. J Med Chem. 2018;61(15):6685–704.
Zhou B, Hu J, Xu F, Chen Z, Bai L, Fernandez-Salas E, et al. Discovery of a small-molecule degrader of bromodomain and extra-terminal (BET) proteins with picomolar cellular potencies and capable of achieving tumor regression. J Med Chem. 2018;61(2):462–81.
Pillow TH, Adhikari P, Blake RA, Chen J, Del Rosario G, Deshmukh G, et al. Antibody conjugation of a chimeric BET degrader enables in vivo activity. ChemMedChem. 2020;15(1):17–25.
Li Y, Yang J, Aguilar A, McEachern D, Przybranowski S, Liu L, et al. Discovery of MD-224 as a first-in-class, highly potent, and efficacious proteolysis targeting chimera murine double minute 2 degrader capable of achieving complete and durable tumor regression. J Med Chem. 2019;62(2):448–66.
Fu S, Wang Y, Keyomarsi K, Meric-Bernstein F. Strategic development of AZD1775, a Wee1 kinase inhibitor, for cancer therapy. Expert Opin Investig Drugs. 2018;27(9):741–51.
Li Z, Pinch BJ, Olson CM, Donovan KA, Nowak RP, Mills CE, et al. Development and characterization of a Wee1 kinase degrader. Cell Chem Biol. 2020;27(1):57-65.e9.
Khan S, Zhang X, Lv D, Zhang Q, He Y, Zhang P, et al. A selective BCL-XL PROTAC degrader achieves safe and potent antitumor activity. Nat Med. 2019;25(12):1938–47.
Winter GE, Mayer A, Buckley DL, Erb MA, Roderick JE, Vittori S, et al. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol Cell. 2017;67(1):5-18.e19.
Woyach JA, Ruppert AS, Guinn D, Lehman A, Blachly JS, Lozanski A, et al. BTKC481S-mediated resistance to ibrutinib in chronic lymphocytic leukemia. J Clin Oncol. 2017;35(13):1437–43.
Chen JG, Liu X, Munshi M, Xu L, Tsakmaklis N, Demos MG, et al. BTK Cys481Ser drives ibrutinib resistance via ERK1/2 and protects BTK wild-type MYD88-mutated cells by a paracrine mechanism. Blood. 2018;131(18):2047–59.
Sun Y, Zhao X, Ding N, Gao H, Wu Y, Yang Y, et al. PROTAC-induced BTK degradation as a novel therapy for mutated BTK C481S induced ibrutinib-resistant B-cell malignancies. Cell Res. 2018;28(7):779–81.
Chessum NEA, Sharp SY, Caldwell JJ, Pasqua AE, Wilding B, Colombano G, et al. Demonstrating in-cell target engagement using a pirin protein degradation probe (CCT367766). J Med Chem. 2018;61(3):918–33.
Brand M, Winter GE. Stick it to E3s. Nat Chem Biol. 2019;15:655–6.
Buckley DL, Raina K, Darricarrere N, Hines J, Gustafson JL, Smith IE, et al. HaloPROTACS: use of small molecule PROTACs to Induce degradation of HaloTag fusion proteins. ACS Chem Biol. 2015;10:1831–7.
Gabizon R, Shraga A, Gehrtz P, Livnah E, Shorer Y, Gurwicz N, et al. Efficient targeted degradation via reversible and irreversible covalent PROTACs. J Am Chem Soc. 2020;142(27):11734–42.
Lu G, Middleton RE, Sun H, Naniong MV, Ott CJ, Mitsiades CS, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of ikaros proteins. Science. 2014;343(6168):305–9.
An J, Ponthier CM, Sack R, Seebacher J, Stadler MB, Donovan KA, et al. PSILAC mass spectrometry reveals ZFP91 as IMiD-dependent substrate of the CRL4 CRBN ubiquitin ligase. Nat Commun. 2017;8:15398.
Ottis P, Palladino C, Thienger P, Britschgi A, Heichinger C, Berrera M, et al. Cellular resistance mechanisms to targeted protein degradation converge toward impairment of the engaged ubiquitin transfer pathway. ACS Chem Biol. 2019;14(10):2215–23.
Zhang L, Riley-Gillis B, Vijay P, Shen Y. Acquired resistance to BET-ProTACS (proteolysis-targeting chimeras) caused by genomic alterations in core components of E3 ligase complexes. Mol Cancer Ther. 2019;18(7):1302–11.
George AJ, Hoffiz YC, Charles AJ, Zhu Y, Mabb AM. A comprehensive atlas of E3 ubiquitin ligase mutations in neurological disorders. Front Genet. 2018;9:29.
Ward CC, Kleinman JI, Brittain SM, Lee PS, Chung CYS, Kim K, et al. Covalent ligand screening uncovers a RNF4 E3 ligase recruiter for targeted protein degradation applications. ACS Chem Biol. 2019;14:2430–40.
Spradlin JN, Hu X, Ward CC, Brittain SM, Jones MD, Ou L, et al. Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat Chem Biol. 2019;15:747–55.
Luo M, Spradlin JN, Brittain SM, McKenna JM, Tallarico JA, Schirle M, et al. Chemoproteomics-enabled ligand screening yields covalent RNF114-based degraders that mimic natural product function. bioRxiv. 2020.
Jiang Y, Deng Q, Zhao H, Xie M, Chen L, Yin F, et al. Development of stabilized peptide-based PROTACs against estrogen receptor α. ACS Chem Biol. 2018;13(3):628–35.
Yamazoe S, Tom J, Fu Y, Wu W, Zeng L, Sun C, et al. Heterobifunctional molecules induce dephosphorylation of kinases-a proof of concept study. J Med Chem. 2020;63(6):2807–13.
Guo WH, Qi X, Yu X, Liu Y, Chung CI, Bai F, et al. Enhancing intracellular accumulation and target engagement of PROTACs with reversible covalent chemistry. Nat Commun. 2020;11(1):4268.
Asatsuma-Okumura T, Ito T, Handa H. Molecular mechanisms of cereblon-based drugs. Pharmacol Ther. 2019;202:132–9.
Neklesa TK, Crews CM. Greasy tags for protein removal. Nature. 2012;487(7407):308–9.
Takahashi D, Moriyama J, Nakamura T, Miki E, Takahashi E, Sato A, et al. AUTACs: cargo-specific degraders using selective autophagy. Mol Cell. 2019;76(5):797-810.e10.
Banik SM, Pedram K, Wisnovsky S, Ahn G, Riley NM, Bertozzi CR. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature. 2020;584(7820):291–7.
Li Z, Zhu C, Ding Y, Fei Y, Lu B. ATTEC: a potential new approach to target proteinopathies. Autophagy. 2020;16(1):185–7.
Acknowledgements
Editorial assistance was provided by the Moffitt Cancer Center’s Office of Scientific Writing by Dr. Paul Fletcher and Daley Drucker. No compensation was given beyond their regular salaries.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
A support grant was received by Dr. Hao Xie from H. Lee Moffitt Cancer Center & Research Institute.
Conflict of interest
The authors, Xinrui Yang, He Yin, Richard D. Kim, Jason B. Fleming, and Hao Xie, declare that they have no conflicts of interest that might be relevant to the contents of this article.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Authors’ contributions
XY contributed to formulating the idea for the manuscript and writing and editing. HX contributed to formulating the idea for the manuscript and editing. HY, RDK, and JBF participated in editing the manuscript.
Rights and permissions
About this article
Cite this article
Yang, X., Yin, H., Kim, R.D. et al. Preclinical and Clinical Advances of Targeted Protein Degradation as a Novel Cancer Therapeutic Strategy: An Oncologist Perspective. Targ Oncol 16, 1–12 (2021). https://doi.org/10.1007/s11523-020-00782-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11523-020-00782-2