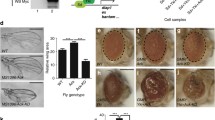
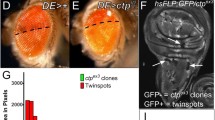
Abstract
The evolutionarily conserved Hippo pathway coordinates cell proliferation, differentiation and apoptosis to regulate organ growth and tumorigenesis. Hippo signaling activity is tightly controlled by various upstream signals including growth factors and cell polarity, but the full extent to which the pathway is regulated during development remains to be resolved. Here, we report the identification of Shaggy, the homolog of mammalian Gsk3β, as a novel regulator of the Hippo pathway in Drosophila. Our results show that Shaggy promotes the expression of Hippo target genes in a manner that is dependent on its kinase activity. Loss of Shaggy leads to Yorkie inhibition and downregulation of Hippo pathway target genes. Mechanistically, Shaggy acts upstream of the Hippo pathway and negatively regulates the abundance of the FERM domain containing adaptor protein Expanded. Our results reveal that Shaggy is functionally required for Crumbs/Slmb-mediated downregulation of Expanded in vivo, providing a potential molecular link between cellular architecture and the Hippo signaling pathway.
Similar content being viewed by others
References
Azzolin, L., Zanconato, F., Bresolin, S., Forcato, M., Basso, G., Bicciato, S., Cordenonsi, M., and Piccolo, S. (2012). Role of TAZ as mediator of Wnt signaling. Cell 151, 1443–1456.
Badouel, C., Gardano, L., Amin, N., Garg, A., Rosenfeld, R., Le Bihan, T., and McNeill, H. (2009). The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell 16, 411–420.
Baumgartner, R., Poernbacher, I., Buser, N., Hafen, E., and Stocker, H. (2010). The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell 18, 309–316.
Bennett, F.C., and Harvey, K.F. (2006). Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol 16, 2101–2110.
Bourouis, M. (2002). Targeted increase in shaggy activity levels blocks wingless signaling. genesis 34, 99–102.
Chen, C.L., Gajewski, K.M., Hamaratoglu, F., Bossuyt, W., Sansores-Garcia, L., Tao, C., and Halder, G. (2010). The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci USA 107, 15810–15815.
Cho, E., Feng, Y., Rauskolb, C., Maitra, S., Fehon, R., and Irvine, K.D. (2006). Delineation of a Fat tumor suppressor pathway. Nat Genet 38, 1142–1150.
Chung, H.L., Augustine, G.J., and Choi, K.W. (2016). Drosophila Schip1 links expanded and Tao-1 to regulate Hippo signaling. Dev Cell 36, 511–524.
Dajani, R., Fraser, E., Roe, S.M., Young, N., Good, V., Dale, T.C., and Pearl, L.H. (2001). Crystal structure of glycogen synthase kinase 3β. Cell 105, 721–732.
Dong, J., Feldmann, G., Huang, J., Wu, S., Zhang, N., Comerford, S.A., Gayyed, M.F., Anders, R.A., Maitra, A., and Pan, D. (2007). Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133.
Duda, P., Akula, S.M., Abrams, S.L., Steelman, L.S., Martelli, A.M., Cocco, L., Ratti, S., Candido, S., Libra, M., Montalto, G., et al. (2020). Targeting GSK3 and associated signaling pathways involved in cancer. Cells 9, 1110.
Feng, Y., and Irvine, K.D. (2007). Fat and expanded act in parallel to regulate growth through warts. Proc Natl Acad Sci USA 104, 20362–20367.
Fiol, C.J., Mahrenholz, A.M., Wang, Y., Roeske, R.W., and Roach, P.J. (1987). Formation of protein kinase recognition sites by covalent modification of the substrate. Molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J Biol Chem 262, 14042–14048.
Frame, S., and Cohen, P. (2001). GSK3 takes centre stage more than 20 years after its discovery. Biochem J 359, 1–16.
Frame, S., Cohen, P., and Biondi, R.M. (2001). A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol Cell 7, 1321–1327.
Fulford, A.D., Holder, M.V., Frith, D., Snijders, A.P., Tapon, N., and Ribeiro, P.S. (2019). Casein kinase 1 family proteins promote Slimb-dependent Expanded degradation. eLife 8, e46592.
Galletti, M., Riccardo, S., Parisi, F., Lora, C., Saqcena, M.K., Rivas, L., Wong, B., Serra, A., Serras, F., Grifoni, D., et al. (2009). Identification of domains responsible for ubiquitin-dependent degradation of dMyc by glycogen synthase kinase 3β and casein kinase 1 kinases. Mol Cell Biol 29, 3424–3434.
Gaspar, P., and Tapon, N. (2014). Sensing the local environment: actin architecture and Hippo signalling. Curr Opin Cell Biol 31, 74–83.
Genevet, A., Wehr, M.C., Brain, R., Thompson, B.J., and Tapon, N. (2010). Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell 18, 300–308.
Grzeschik, N.A., Parsons, L.M., Allott, M.L., Harvey, K.F., and Richardson, H.E. (2010). Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol 20, 573–581.
Hafezi, Y., Bosch, J.A., and Hariharan, I.K. (2012). Differences in levels of the transmembrane protein Crumbs can influence cell survival at clonal boundaries. Dev Biol 368, 358–369.
Halder, G., and Johnson, R.L. (2011). Hippo signaling: growth control and beyond. Development 138, 9–22.
Hart, M., Concordet, J.P., Lassot, I., Albert, I., del los Santos, R., Durand, H., Perret, C., Rubinfeld, B., Margottin, F., Benarous, R., et al. (1999). The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr Biol 9, 207–211.
Huang, J., Wu, S., Barrera, J., Matthews, K., and Pan, D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421–434.
Huang, W., Lv, X., Liu, C., Zha, Z., Zhang, H., Jiang, Y., Xiong, Y., Lei, Q. Y., and Guan, K.L. (2012). The N-terminal phosphodegron targets TAZ/WWTR1 protein for SCFβ-TrCP-dependent degradation in response to phosphatidylinositol 3-kinase inhibition. J Biol Chem 287, 26245–26253.
Kanuka, H., Kuranaga, E., Takemoto, K., Hiratou, T., Okano, H., and Miura, M. (2005). Drosophila caspase transduces Shaggy/GSK-3β kinase activity in neural precursor development. EMBO J 24, 3793–3806.
Karaman, R., and Halder, G. (2018). Cell junctions in Hippo signaling. Cold Spring Harb Perspect Biol 10, a028753.
Ling, C., Zheng, Y., Yin, F., Yu, J., Huang, J., Hong, Y., Wu, S., and Pan, D. (2010). The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci USA 107, 10532–10537.
Liu, C., Kato, Y., Zhang, Z., Do, V.M., Yankner, B.A., and He, X. (1999). β-Trcp couples β-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc Natl Acad Sci USA 96, 6273–6278.
Liu, C., Li, Y., Semenov, M., Han, C., Baeg, G.H., Tan, Y., Zhang, Z., Lin, X., and He, X. (2002). Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108, 837–847.
Ma, X., Guo, X., Richardson, H.E., Xu, T., and Xue, L. (2018). POSH regulates Hippo signaling through ubiquitin-mediated expanded degradation. Proc Natl Acad Sci USA 115, 2150–2155.
Martinek, S., Inonog, S., Manoukian, A.S., and Young, M.W. (2001). A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105, 769–779.
McCartney, B.M., Kulikauskas, R.M., LaJeunesse, D.R., and Fehon, R.G. (2000). The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development 127, 1315–1324.
Misra, J.R., and Irvine, K.D. (2018). The Hippo signaling network and its biological functions. Annu Rev Genet 52, 65–87.
Oh, H., and Irvine, K.D. (2008). In vivo regulation of Yorkie phosphorylation and localization. Development 135, 1081–1088.
Oh, H., and Irvine, K.D. (2009). In vivo analysis of Yorkie phosphorylation sites. Oncogene 28, 1916–1927.
Oh, H., Reddy, B.V.V.G., and Irvine, K.D. (2009). Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev Biol 335, 188–197.
Orme, M.H., Liccardi, G., Moderau, N., Feltham, R., Wicky-John, S., Tenev, T., Aram, L., Wilson, R., Bianchi, K., Morris, O., et al. (2016). The unconventional myosin CRINKLED and its mammalian orthologue MYO7A regulate caspases in their signalling roles. Nat Commun 7, 10972.
Panciera, T., Azzolin, L., Cordenonsi, M., and Piccolo, S. (2017). Mechanobiology of YAP and TAZ in physiology and disease. Nat Rev Mol Cell Biol 18, 758–770.
Papadopoulou, D., Bianchi, M.W., and Bourouis, M. (2004). Functional studies of Shaggy/Glycogen Synthase Kinase 3 phosphorylation sites in Drosophila melanogaster. Mol Cell Biol 24, 4909–4919.
Patel, P., and Woodgett, J.R. (2017). Glycogen synthase kinase 3: a kinase for all pathways? Curr Top Dev Biol 123, 277–302.
Penzo-Méndez, A.I., and Stanger, B.Z. (2015). Organ-size regulation in mammals. Cold Spring Harb Perspect Biol 7, a019240.
Price, M.A., and Kalderon, D. (2002). Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell 108, 823–835.
Rauskolb, C., Pan, G., Reddy, B.V.V.G., Oh, H., and Irvine, K.D. (2011). Zyxin links fat signaling to the hippo pathway. PLoS Biol 9, e1000624.
Ribeiro, P., Holder, M., Frith, D., Snijders, A.P., and Tapon, N. (2014). Crumbs promotes expanded recognition and degradation by the SCFSlimb/β-TrCP ubiquitin ligase. Proc Natl Acad Sci USA 111, E1980–E1989.
Robertson, H., Hayes, J.D., and Sutherland, C. (2018). A partnership with the proteasome; the destructive nature of GSK3. Biochem Pharmacol 147, 77–92.
Robinson, B.S., Huang, J., Hong, Y., and Moberg, K.H. (2010). Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol 20, 582–590.
Ruel, L., Bourouis, M., Heitzler, P., Pantesco, V., and Simpson, P. (1993a). Drosophila shaggy kinase and rat glycogen synthase kinase-3 have conserved activities and act downstream of Notch. Nature 362, 557–560.
Ruel, L., Pantesco, V., Lutz, Y., Simpson, P., and Bourouis, M. (1993b). Functional significance of a family of protein kinases encoded at the shaggy locus in Drosophila. EMBO J 12, 1657–1669.
Sears, R., Nuckolls, F., Haura, E., Taya, Y., Tamai, K., and Nevins, J.R. (2000). Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14, 2501–2514.
Silva, E., Tsatskis, Y., Gardano, L., Tapon, N., and McNeill, H. (2006). The tumor-suppressor gene fat controls tissue growth upstream of expanded in the Hippo signaling pathway. Curr Biol 16, 2081–2089.
Smelkinson, M.G., and Kalderon, D. (2006). Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component Slimb. Curr Biol 16, 110–116.
Song, Z., Han, X., Zou, H., Zhang, B., Ding, Y., Xu, X., Zeng, J., Liu, J., and Gong, A. (2018). PTEN-GSK3β-MOB1 axis controls neurite outgrowth in vitro and in vivo. Cell Mol Life Sci 75, 4445–4464.
Su, T., Ludwig, M.Z., Xu, J., and Fehon, R.G. (2017). Kibra and Merlin activate the Hippo pathway spatially distinct from and independent of expanded. Dev Cell 40, 478–490.e3.
Sutherland, C. (2011). What are the bona fide GSK3 substrates? Int J Alzheimers Dis 2011, 1–23.
Tumaneng, K., Russell, R.C., and Guan, K.L. (2012). Organ size control by Hippo and TOR pathways. Curr Biol 22, R368–R379.
Vrabioiu, A.M., and Struhl, G. (2015). Fat/Dachsous signaling promotes Drosophila wing growth by regulating the conformational state of the NDR kinase warts. Dev Cell 35, 737–749.
Wang, X., Zhang, Y., and Blair, S.S. (2019). Fat-regulated adaptor protein Dlish binds the growth suppressor Expanded and controls its stability and ubiquitination. Proc Natl Acad Sci USA 116, 1319–1324.
Willecke, M., Hamaratoglu, F., Kango-Singh, M., Udan, R., Chen, C.L., Tao, C., Zhang, X., and Halder, G. (2006). The fat cadherin acts through the Hippo tumor-suppressor pathway to regulate tissue size. Curr Biol 16, 2090–2100.
Yanagawa, S., Matsuda, Y., Lee, J.S., Matsubayashi, H., Sese, S., Kadowaki, T., and Ishimoto, A. (2002). Casein kinase I phosphorylates the Armadillo protein and induces its degradation in Drosophila. EMBO J 21, 1733–1742.
Yang, S., Xu, W., Liu, C., Jin, J., Li, X., Jiang, Y., Zhang, L., Meng, X., Zhan, J., and Zhang, H. (2022). LATS1 K751 acetylation blocks activation of Hippo signalling and switches LATS1 from a tumor suppressor to an oncoprotein. Sci China Life Sci 65, 129–141.
Yost, C., Torres, M., Miller, J.R., Huang, E., Kimelman, D., and Moon, R.T. (1996). The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev 10, 1443–1454.
Yu, F.X., and Guan, K.L. (2013). The Hippo pathway: regulators and regulations. Genes Dev 27, 355–371.
Yu, F.X., Zhao, B., and Guan, K.L. (2015). Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811–828.
Yu, J., Zheng, Y., Dong, J., Klusza, S., Deng, W.M., and Pan, D. (2010). Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell 18, 288–299.
Zhang, H., Li, C., Chen, H., Wei, C., Dai, F., Wu, H., Dui, W., Deng, W.M., and Jiao, R. (2015). SCFSlmb E3 ligase-mediated degradation of Expanded is inhibited by the Hippo pathway in Drosophila. Cell Res 25, 93–109.
Zhang, Q., Lin, F., Huang, J., and Xiong, C. (2022). Mechanical transmission enables EMT cancer cells to drive epithelial cancer cell migration to guide tumor spheroid disaggregation. Sci China Life Sci, doi: https://doi.org/10.1007/s11427-021-2054-3.
Zheng, Y., and Pan, D. (2019). The Hippo signaling pathway in development and disease. Dev Cell 50, 264–282.
Acknowledgements
This study was supported by the National Key R&D Program of China (2021YFA0805800, 2020YFA0803202, 2018YFC1003203, 2021YFC2700403), the National Natural Science Foundation of China (31970538, 32000574, 31871452), the Guangzhou Medical University Discipline Construction Funds (Basic Medicine) (JCXKJS2022A02), the 111 Project (D18010), the Local Innovative and Research Teams Project of Guangdong Perl River Talents Program (2017BT01S155), the Special Innovation Projects of Universities in Guangdong Province (2018KTSCX182), the Medical Scientific Research Foundation of Guangdong Province (A2019292), and the Natural Science Foundation of Guangdong Province (2017A030310403). We are grateful to Drs. Duojia Pan, Richard Fehon, Lei Zhang and Daniel Kalderon for kindly sharing reagents. We acknowledge members of the Jiao laboratory for discussions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Compliance and ethics All author(s) declare that they have no conflict of interest.
Supplementary Figures
Rights and permissions
About this article
Cite this article
Wu, H., Zhu, N., Liu, J. et al. Shaggy regulates tissue growth through Hippo pathway in Drosophila. Sci. China Life Sci. 65, 2131–2144 (2022). https://doi.org/10.1007/s11427-022-2156-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-022-2156-2