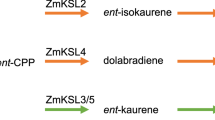
Abstract
Land plants co-speciate with a diversity of continually expanding plant specialized metabolites (PSMs) and root microbial communities (microbiota). Homeostatic interactions between plants and root microbiota are essential for plant survival in natural environments. A growing appreciation of microbiota for plant health is fuelling rapid advances in genetic mechanisms of controlling microbiota by host plants. PSMs have long been proposed to mediate plant and single microbe interactions. However, the effects of PSMs, especially those evolutionarily new PSMs, on root microbiota at community level remain to be elucidated. Here, we discovered sesterterpenes in Arabidopsis thaliana, produced by recently duplicated prenyltransferase-terpene synthase (PT-TPS) gene clusters, with neo-functionalization. A single-residue substitution played a critical role in the acquisition of sesterterpene synthase (sesterTPS) activity in Brassicaceae plants. Moreover, we found that the absence of two root-specific sesterterpenoids, with similar chemical structure, significantly affected root microbiota assembly in similar patterns. Our results not only demonstrate the sensitivity of plant microbiota to PSMs but also establish a complete framework of host plants to control root microbiota composition through evolutionarily dynamic PSMs.
Similar content being viewed by others
References
Aubourg, S., Lecharny, A., and Bohlmann, J. (2002). Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genets Genom 267, 730–745.
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc-Ser B 57, 289–300.
Berendsen, R.L., Pieterse, C.M.J., and Bakker, P.A.H.M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci 17, 478–486.
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., Rauf, P., Huettel, B., Reinhardt, R., Schmelzer, E., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95.
Caporaso, J.G., Bittinger, K., Bushman, F.D., DeSantis, T.Z., Andersen, G. L., and Knight, R. (2010a). PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267.
Caporaso, J.G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F.D., Costello, E.K., Fierer, N., Peña, A.G., Goodrich, J.K., Gordon, J.I., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336.
Castrillo, G., Teixeira, P.J.P.L., Paredes, S.H., Law, T.F., de Lorenzo, L., Feltcher, M.E., Finkel, O.M., Breakfield, N.W., Mieczkowski, P., Jones, C.D., et al. (2017). Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518.
Chen, F., Tholl, D., Bohlmann, J., and Pichersky, E. (2011). The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66, 212–229.
Chen, H., and Boutros, P.C. (2011). VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinf 12, 35.
Chen, Q., Fan, D., and Wang, G. (2015). Heteromeric geranyl (geranyl) diphosphate synthase is involved in monoterpene biosynthesis in Arabidopsis flowers. Mol Plant 8, 1434–1437.
Christianson, D.W. (2017). Structural and chemical biology of terpenoid cyclases. Chem Rev 117, 11570–11648.
DeSantis, T.Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E.L., Keller, K., Huber, T., Dalevi, D., Hu, P., and Andersen, G.L. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72, 5069–5072.
Edgar, R.C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461.
Edgar, R.C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10, 996–998.
Edgar, R.C., Haas, B.J., Clemente, J.C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200.
Foster, K.R., Schluter, J., Coyte, K.Z., and Rakoff-Nahoum, S. (2017). The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51.
Gan, X., Hay, A., Kwantes, M., Haberer, G., Hallab, A., Ioio, R.D., Hofhuis, H., Pieper, B., Cartolano, M., Neumann, U., et al. (2016). The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat Plants 2, 16167.
Hartmann, T. (2007). From waste products to ecochemicals: Fifty years research of plant secondary metabolism. Phytochemistry 68, 2831–2846.
Huang, A.C., Kautsar, S.A., Hong, Y.J., Medema, M.H., Bond, A.D., Tantillo, D.J., and Osbourn, A. (2017). Unearthing a sesterterpene biosynthetic repertoire in the Brassicaceae through genome mining reveals convergent evolution. Proc Natl Acad Sci USA 114, E6005–E6014.
Kampranis, S.C., Ioannidis, D., Purvis, A., Mahrez, W., Ninga, E., Katerelos, N.A., Anssour, S., Dunwell, J.M., Degenhardt, J., Makris, A. M., et al. (2007). Rational conversion of substrate and product specificity in a Salvia monoterpene synthase: Structural insights into the evolution of terpene synthase function. Plant Cell 19, 1994–2005.
Katoh, K., Misawa, K., and Kuma, K.I. (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res 30, 3059–3066.
Kliebenstein, D.J., and Osbourn, A. (2012). Making new molecules—Evolution of pathways for novel metabolites in plants. Curr Opin Plant Biol 15, 415–423.
Leach, J.E., Triplett, L.R., Argueso, C.T., and Trivedi, P. (2017). Communication in the phytobiome. Cell 169, 587–596.
Lebeis, S.L., Paredes, S.H., Lundberg, D.S., Breakfield, N., Gehring, J., McDonald, M., Malfatti, S., Glavina del Rio, T., Jones, C.D., Tringe, S. G., et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864.
Li, W., Zhang, F., Chang, Y., Zhao, T., Schranz, M.E., and Wang, G. (2015). Nicotinate O-glucosylation is an evolutionarily metabolic trait important for seed germination under stress conditions in Arabidopsis thaliana. Plant Cell 21, 1907–1924.
Lundberg, D.S., Lebeis, S.L., Paredes, S.H., Yourstone, S., Gehring, J., Malfatti, S., Tremblay, J., Engelbrektson, A., Kunin, V., Del Rio, T.G., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90.
Müller, D.B., Vogel, C., Bai, Y., and Vorholt, J.A. (2016). The plant microbiota: Systems-level insights and perspectives. Annu Rev Genet 50, 211–234.
Murrell, B., Wertheim, J.O., Moola, S., Weighill, T., Scheffler, K., and Kosakovsky Pond, S.L. (2012). Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8, e1002764.
Robinson, M.D., McCarthy, D.J., and Smyth, G.K. (2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140.
Schlaeppi, K., Dombrowski, N., Oter, R.G., Ver Loren van Themaat, E., and Schulze-Lefert, P. (2014). Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci USA 111, 585–592.
Shao, J., Chen, Q.W., Lv, H.J., He, J., Liu, Z.F., Lu, Y.N., Liu, H.L., Wang, G.D., and Wang, Y. (2011). (+)-Thalianatriene and (-)-retigeranin B catalyzed by sesterterpene synthases from Arabidopsis thaliana. Org Lett 19, 1816–1819.
Srividya, N., Davis, E.M., Croteau, R.B., and Markus Lange, B. (2015). Functional analysis of (4S)-limonene synthase mutants reveals determinants of catalytic outcome in a model monoterpene synthase. Proc Natl Acad Sci USA 112, 3332–3337.
Starks, C.M., Back, K., and Chappell, J. (1997). Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science 277, 1815–1820.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis Version 6.0. Mol Biol Evol 30, 2725–2729.
Tholl, D., and Lee, S. (2011). Terpene specialized metabolism in Arabidopsis thaliana. Arabidopsis Book 9, e0143.
van Dam, N.M., and Bouwmeester, H.J. (2016). Metabolomics in the rhizosphere: Tapping into belowground chemical communication. Trends Plant Sci 21, 256–265.
Verbon, E.H., and Liberman, L.M. (2016). Beneficial microbes affect endogenous mechanisms controlling root development. Trends Plant Sci 21, 218–229.
Vickers, C.E., Bongers, M., Liu, Q., Delatte, T., and Bouwmeester, H. (2014). Metabolic engineering of volatile isoprenoids in plants and microbes. Plant Cell Environ 37, 1753–1775.
Wang, C., Chen, Q., Fan, D., Li, J., Wang, G., and Zhang, P. (2016). Structural analyses of short-chain prenyltransferases identify an evolutionarily conserved GFPPS clade in Brassicaceae plants. Mol Plant 9, 195–204.
Wang, G., Tian, L., Aziz, N., Broun, P., Dai, X., He, J., King, A., Zhao, P. X., and Dixon, R.A. (2008). Terpene biosynthesis in glandular trichomes of hop. Plant Physiol 148, 1254–1266.
Wang, Q., Garrity, G.M., Tiedje, J.M., and Cole, J.R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ MicroBiol 73, 5261–5267.
Xu, H., Zhang, F., Liu, B., Huhman, D.V., Sumner, L.W., Dixon, R.A., and Wang, G. (2013). Characterization of the formation of branched short-chain fatty acid: CoAs for bitter acid biosynthesis in hop glandular trichomes. Mol Plant 6, 1301–1317.
Zhang, J., Zhang, N., Liu, Y.X., Zhang, X., Hu, B., Qin, Y., Xu, H., Wang, H., Guo, X., Qian, J., et al. (2018). Root microbiota shift in rice correlates with resident time in the field and developmental stage. Sci China Life Sci 61, 613–621.
Zhao, T., Holmer, R., de Bruijn, S., Angenent, G.C., van den Burg, H.A., and Schranz, M.E. (2011). Phylogenomic synteny network analysis of MADS-box transcription factor genes reveals lineage-specific transpositions, ancient tandem duplications, and deep positional conservation. Plant Cell 29, 1278–1292.
Acknowledgements
This work was supported by the Priority Research Program of the Chinese Academy of Sciences (ZDRW-ZS-2019-2 and QYZDB-SSW-SMC021), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA08000000 and XDB11020700), the National Program on Key Basic Research Projects (2013CB127000), and the State Key Laboratory of Plant Genomics of China (2016A0219-11 and SKLPG2013A0125-5). We thank Dr. Jay D Keasling (University of California, Berkeley) for providing the pMBIS plasmid.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supporting Information
Figure S1 Functional characterization of 16 plant sesterTPSs in E.coli system.
Figure S2 Terpene synthase assays of TPS18 and TPS18G328W analyzed by GC-MS.
Figure S3 Terpene synthase assays of TPS19 and TPS19G325W analyzed by GC-MS.
Figure S4 Terpene synthase assays of TPS25 and TPS25G328W analyzed by GC-MS.
Figure S5 Terpene synthase assays of TPS30 and TPS30P328W analyzed by GC-MS.
Figure S6 Characterization of TPS25 T-DNA insertion mutants.
Figure S7 Characterization of TPS30 T-DNA insertion mutant (tps30-1|CS805958).
Figure S8tps30-2 and tps30/tps25-1 double mutants generated via CRISPR/Cas9.
Figure S9tps30-3 and tps30/tps25-2 double mutants generated via CRISPR/Cas9.
Figure S10 Sesterterpene profiling in various tissues of Arabidopsis and different transgenic plants by GC-QQQ-MS.
Figure S11 Phenotypes of sesterTPS mutants and Col-0 when grown in natural soil (Changping farm in Beijing) under climate control conditions.
Figure S12 Sample diversity (α/β-diversity) measurements among each genotype.
Figure S13 Stack plot showing relative abundance distribution of the OTUs in phylum.
Figure S14 Taxonomic difference and overlap of specific differential OTUs between TPS mutants and Col-0.
Table S1 The MRM settings for four sesterterpene (C25) backbones analyzed by GC-QQQ-MS in this study
Table S2 The primers used in this study
The supporting information is available online at http://life.scichina.com and https://link.springer.com. The supporting materials are published as submitted, without typesetting or editing. The responsibility for scientific accuracy and content remains entirely with the authors.
Supplemental Information
Rights and permissions
About this article
Cite this article
Chen, Q., Jiang, T., Liu, YX. et al. Recently duplicated sesterterpene (C25) gene clusters in Arabidopsis thaliana modulate root microbiota. Sci. China Life Sci. 62, 947–958 (2019). https://doi.org/10.1007/s11427-019-9521-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-019-9521-2