Abstract
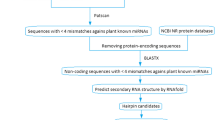
microRNAs (miRNAs) are a class of endogenous, non-coding, short (∼21 nt) RNAs directly involved in regulating gene expression at the post-transcriptional level. Previous reports have noted that plant miRNAs in the plant kingdom are highly conserved, which provides the foundation for identification of conserved miRNAs in other plant species through homology alignment. Conserved miRNAs in wheat are identified using EST (Expressed Sequence Tags) and GSS analysis. All previously known miRNAs in other plant species were blasted against wheat EST and GSS sequences to select novel miRNAs in wheat by a series of filtering criteria. From a total of 37 conserved miRNAs belonging to 18 miRNA families 10 conserved miRNAs comprising 4 families were reported in wheat. MiR395 is found to be a special family, because three members belonging to the same miR395 family are clustered together, similar to animal miRNAs. MiRNA targets are transcription factors involved in wheat growth and development, metabolism,and stress responses.
Similar content being viewed by others
References
Bartel D P. microRNAs: Genomics, biogenesis, mechanism, and function. Cell, 2004, 116: 281–297, 10.1016/S0092-8674(04)00045-5, 1:CAS:528:DC%2BD2cXhtVals7o%3D, 14744438
Jones-Rhoades M W, Bartel D P, Bartel B. microRNAs and their regulatory roles in plants. Annu Rev Plant Biol, 2006, 57: 19–53, 10.1146/annurev.arplant.57.032905.105218, 1:CAS:528:DC%2BD28XosVKhsb0%3D, 16669754
Papp I, Mette M F, Aufsatz W, et al. Evidence for nuclear processing plant microRNA and short interfering precursors. Plant Physiol, 2003, 132: 1382–1390, 10.1104/pp.103.021980, 1:CAS:528:DC%2BD3sXlsFGhtbg%3D, 12857820
Park M Y, Wu G, Gonzalez-Sulser A, et al. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acid Sci USA, 2005, 102: 3691–3696, 10.1073/pnas.0405570102, 1:CAS:528:DC%2BD2MXisVOgsbw%3D
Arazi T, Talmor-Neiman M, Stav R, et al. Cloning and characterization of microRNAs from moss. Plant J, 2005, 43: 837–848, 10.1111/j.1365-313X.2005.02499.x, 1:CAS:528:DC%2BD2MXhtVKqtrvK, 16146523
Sunkar R, Zhu J K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell, 2004, 16: 2001–2019, 10.1105/tpc.104.022830, 1:CAS:528:DC%2BD2cXmvVansb0%3D, 15258262
Sunkar R, Girke T, Jain P K, et al. Cloning and characterization of microRNAs from rice. Plant Cell, 2005, 17: 1397–1411, 10.1105/tpc.105.031682, 1:CAS:528:DC%2BD2MXksVKksrc%3D, 15805478
Zhang B H, Pan X P, Cannon C H, et al. Conservation and divergence of plant microRNA genes. Plant J, 2006, 46: 243–259, 10.1111/j.1365-313X.2006.02697.x, 1:CAS:528:DC%2BD28XksVWntrk%3D, 16623887
Lee R C, Feinbaum R L, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 1993, 75: 843–854, 10.1016/0092-8674(93)90529-Y, 1:CAS:528:DyaK2cXpslGqtA%3D%3D, 8252621
Brown J R, Sanseau P. A computational view of microRNAs and their targets. Drug Discov Today, 2005, 10: 595–601, 10.1016/S1359-6446(05)03399-4, 1:CAS:528:DC%2BD2MXjtlahtL4%3D, 15837603
Lu S, Sun Y H, Shi R, et al. Novel and mechanical stress-responsive micrornas in Populus trichocarpa that are absent from Arabidopsis. Plant Cell, 2005, 17: 2186–2203, 10.1105/tpc.105.033456, 1:CAS:528:DC%2BD2MXpsFGjs7Y%3D, 15994906
Moxon S J, Szittya R C, Schwach G, et al. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res, 2008, 18: 1602–1609, 10.1101/gr.080127.108, 1:CAS:528:DC%2BD1cXht1elu7%2FF, 18653800
Qiu D Y, Pan X P, Wilson W I, et al. High throughput sequencing technology reveals that the taxoid elicitor methyl jasmonate regulates microRNA expression in Chinese yew (Taxus chinensis). Gene, 2009, 436: 37–44, 10.1016/j.gene.2009.01.006, 1:CAS:528:DC%2BD1MXjsVCgsbY%3D, 19393185
Jones-Rhoades M W, Bartel D P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell, 2004, 14: 787–799, 10.1016/j.molcel.2004.05.027, 1:CAS:528:DC%2BD2cXlslemtrs%3D, 15200956
Wang X J, Reyes J L, Chua N H, et al. Prediction and identification of Arabidopsis thaliana microRNAs and their targets. Genome Biol, 2004, 5: R65, 10.1186/gb-2004-5-9-r65, 15345049
Bonnet E, Wuyts J, Rouze P, et al. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important targets. Proc Natl Acad Sci USA, 2004, 101: 11511–11516, 10.1073/pnas.0404025101, 1:CAS:528:DC%2BD2cXmvVKgu7g%3D, 15272084
Zhang B H, Pan X P, Wang Q L, et al. Identification and characterization of now plant microRNAs using EST analysis. Cell Res, 2005, 15: 336–360, 10.1038/sj.cr.7290302, 15916721
Zhang B H, Pan X P, Anderson T A. Identification of 188 conserved maize microRNAs and their targets. FEBS Lett, 2006, 580: 3753–3762, 10.1016/j.febslet.2006.05.063, 1:CAS:528:DC%2BD28XmtVOmurg%3D, 16780841
Qiu C X, Xie F L, Zhu Y Y, et al. Computational identification of microRNAs and their targets in Gossypium hirsutum expressed sequence tags. Gene, 2007, 395: 49–61, 10.1016/j.gene.2007.01.034, 1:CAS:528:DC%2BD2sXkvFWqsr8%3D, 17408884
Jin W, Li N, Zhang B, et al. Identification and verification of microRNA in wheat (Triticum aestivum). J Plant Res, 2008, 121: 351–355, 10.1007/s10265-007-0139-3, 1:CAS:528:DC%2BD1cXls1KltLg%3D, 18357413
Yao Y, Guo G, Ni Z, et al. Cloning and characterization of microRNAs from wheat (Tricitum aestivum L.). Genome Biol, 2007, 8: R96, 10.1186/gb-2007-8-6-r96, 17543110
Dryanova A, Zakharov A, Gulick P J. Data mining for miRNAs and their targets in the Triticeae. Genome, 2008, 51: 433–443, 10.1139/G08-025, 1:CAS:528:DC%2BD1cXmsVOjurk%3D, 18521122
Griffiths-Jones S, Grocock R J, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res, 2006, 34: D140–D144, 10.1093/nar/gkj112, 1:CAS:528:DC%2BD28XisFyhtw%3D%3D, 16381832
Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res, 2003, 31: 3406–3415, 10.1093/nar/gkg595, 1:CAS:528:DC%2BD3sXltVWisr8%3D, 12824337
Meyers B C, Axtell M J, Bartel B, et al. Criteria for annotation of plant microRNAs. Plant Cell, 2008, 20: 3186–3190, 10.1105/tpc.108.064311, 1:CAS:528:DC%2BD1MXitVartbo%3D, 19074682
Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res, 1988, 16: 10881–10890, 10.1093/nar/16.22.10881, 1:CAS:528:DyaL1MXjsVOqtA%3D%3D, 2849754
Zhang B H, Pan X P, Cox B, et al. Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci, 2006, 63: 246–254, 10.1007/s00018-005-5467-7, 1:CAS:528:DC%2BD28Xhs1Gnt7k%3D, 16395542
Yu J, Wang F, Yang G H, et al. Human microRNA clusters: Genomic organization and expression profile in leukemina cell lines. Biochem Biophys Res Commun, 2006, 349: 59–68, 10.1016/j.bbrc.2006.07.207, 1:CAS:528:DC%2BD28XpsVKqsLs%3D, 16934749
Talmor-Neiman M, Stva R, Framk W, et al. Novel microRNAs and intermediates of microRNA biogenesis from moss. Plant J, 2006, 47: 25–37, 10.1111/j.1365-313X.2006.02768.x, 1:CAS:528:DC%2BD28XnvFaltbs%3D, 16824179
Lu S, Sun Y H, Amerson H, et al. MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J, 2007, 51: 1077–1098, 10.1111/j.1365-313X.2007.03208.x, 1:CAS:528:DC%2BD2sXhtFKnur7F, 17635765
Allen E, Xie Z, Gustafson A M, et al. MicroRNA-directed phrasing during trans-acting siRNA biogenesis in plants. Cell, 2005, 121: 207–221, 10.1016/j.cell.2005.04.004, 1:CAS:528:DC%2BD2MXjvV2jtb4%3D, 15851028
Guo H S, Xie Q, Fei J F, et al. microRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell, 2005, 17: 1376–1386, 10.1105/tpc.105.030841, 1:CAS:528:DC%2BD2MXksVKksrk%3D, 15829603
Mallory A C, Bartel D P. microRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulate expression of early auxin response genes. Plant Cell, 2005, 17: 1360–1375, 10.1105/tpc.105.031716, 1:CAS:528:DC%2BD2MXksVKksrg%3D, 15829600
Palatnik J F, Allen E, Wu X, et al. Control of leaf morphogenesis by microRNAs. Nature, 2003, 425: 257–263, 10.1038/nature01958, 1:CAS:528:DC%2BD3sXntlWns7w%3D, 12931144
Lee Y, Kim M, Han J, et al. microRNA genes are transcribed by RNA polymerase II. EMBO J, 2004, 23: 4051–4060, 10.1038/sj.emboj.7600385, 1:CAS:528:DC%2BD2cXotlCrsrs%3D, 15372072
Rhoades M W, Reinhart B J, Lim L P, et al. Prediction of plant microRNA targets. Cell, 2002, 110: 513–520, 10.1016/S0092-8674(02)00863-2, 1:CAS:528:DC%2BD38Xmslyjt7s%3D, 12202040
Bartel B, Bartel D P. microRNAs: at the root of plant development. Plant Physiol, 2003, 132: 709–717, 10.1104/pp.103.023630, 1:CAS:528:DC%2BD3sXkslertro%3D, 12805599
Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science, 2004, 303: 2022–2025, 10.1126/science.1088060, 1:CAS:528:DC%2BD2cXisVGlt7g%3D, 12893888
Aukerman M J, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell, 2003, 15: 2730–2741, 10.1105/tpc.016238, 1:CAS:528:DC%2BD3sXpt1OrurY%3D, 14555699
Park W, Li J, Song R, et al. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Bio, 2002, 12: 1484–1495, 10.1016/S0960-9822(02)01017-5, 1:CAS:528:DC%2BD38XntVaisb4%3D
Mallory A C, Dugas D V, Bartel D P, et al. microRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol, 2004, 14: 1035–1046, 10.1016/j.cub.2004.06.022, 1:CAS:528:DC%2BD2cXltVOgtLg%3D, 15202996
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by the National Natural Science Foundation of China (Grant Nos. 30600421 and 30430490) and the National High Technology Research and Development Program of China (Grant No. 2006BAD22BO1)
Rights and permissions
About this article
Cite this article
Han, Y., Luan, F., Zhu, H. et al. Computational identification of microRNAs and their targets in wheat (Triticum aestivum L.). SCI CHINA SER C 52, 1091–1100 (2009). https://doi.org/10.1007/s11427-009-0144-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-009-0144-y