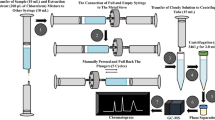
Abstract
In the present study, the uptake and metabolization of the sartan drug telmisartan by a series of plants was investigated. Thereby for seven potential metabolites, modifications on the telmisartan molecule such as hydroxylation and/or glycosylation could be tentatively identified. For two additional signals detected at accurate masses m/z 777.3107 and m/z 793.3096, no suggestions for molecular formulas could be made. Further investigations employing garden cress (Lepidium sativum) as a model plant were conducted. This was done in order to develop an analytical method allowing the detection of these substances also under environmentally relevant conditions. For this reason, the knowledge achieved from treatment of the plants with rather high concentrations of the parent drug (10 mg L−1) was compared with results obtained when using solutions containing telmisartan in the μg - ng L−1 range. Thereby the parent drug and up to three tentative drug-related metabolites could still be detected. Finally cress was cultivated in water taken from a local waste water treatment plant effluent containing 90 ng L−1 of telmisartan and harvested and the cress roots were extracted. In this extract, next to the parent drug one major metabolite, namely telmisartan-glucose could be identified.
Similar content being viewed by others
Introduction
Since the first successful detection of drugs (respectively drug metabolites) in the aquatic system by Hignite and Azarnoff (1977), environmental waters and in particular effluents of wastewater treatment plants (WWTP) have been frequently analyzed with respect to the presence of drug residues (for exemplary reviews, see Buchberger 2011; Cotton et al. 2016; Lindberg et al. 2014; Petrie et al. 2013; Puckowski et al. 2016; Richardson and Ternes 2014; Rzymski et al. 2017). Within the last years, a new facet related to the certainty that pharmaceuticals can be found in the effluent of WWTPs all over the world has emerged, as those waters are increasingly used for irrigation purposes in agriculture. Main reasons for this practice are rising problems with droughts paired with an increasing demand for food and feed (due to the continuous growth of the world´s population) triggering a search for new sources of water (European Commission 2013, 2008). Although such reclaimed waters used in the irrigation of plants intended for human or animal consumption must fulfill certain minimum standards (mainly with respect to microbiological and physico-chemical parameters), until now no regulation focusing on the presence of emerging contaminants (such as pharmaceuticals) exists (Alcalde-Sanz and Gawlik 2017). Furthermore, certain plants have been tested and subsequently employed for their potential in the removal of xenobiotics from wastewater in constructed wetlands (Nuel et al. 2018). All these facts have led to a growing interest in investigations dealing with the interaction between plants and these emerging contaminants added through irrigation water or contaminated fertilizer (e.g., manure from treated animals or sludge from WWTPs) (for relevant reviews, see Fu et al. 2019; Kinney and van den Heuvel 2020; Klampfl 2019; Pico et al. 2017; Reddy Pullagurala et al. 2018). Among these studies, those mainly directed towards investigating the uptake of the parent drug (or its metabolite) from water or soil, and a second type primarily devoted to research on the further biotransformation of the drug focusing on the identification of new metabolites formed within the plant can be distinguished (Klampfl 2019; Klampfl et al. 2021). Thereby, within the first group, the focus was often placed on experimental set-ups that were as much as possible resembling real environmental conditions. Therefore, from an analytical point of view, approaches suitable for ultra-trace analysis of the parent drug and sometimes its main metabolite(s) have to be pursued (for exemplary papers, see Beltrán et al. 2020; Keerthanan et al. 2021; Montemurro et al. 2017; Reddy Pullagurala et al. 2018). When the elucidation of the biological fate of the drug (after uptake) within the plant was the main goal, plants were often treated with much higher concentrations of the drug under investigation to facilitate the detection of as many metabolites as possible (for exemplary papers see, e.g., Emhofer et al. 2019; Klampfl 2019; Mlynek et al. 2020). Here the main focus was set on the tentative identification of structures and the metabolic pathways responsible for the formation of these substances. However, it should be stated that of course a series of research papers not exactly fitting into this scheme exists (e.g., Emhofer et al. 2018; Riemenschneider et al. 2017). Throughout the years, an ever-increasing plentitude of different drugs but also personal care products, summarized as pharmaceuticals and personal care products (PPCPs), was included in these studies. For about five years, the group of sartan drugs, such as valsartan or telmisartan (TEL), has been added to this portfolio with studies on the presence of these substances in environmental waters (Bayer et al. 2014; Castro et al. 2019; Giebułtowicz et al. 2016). The interaction of sartans with plants has so far only been studied in a single paper reporting the uptake and metabolization of this drug by Salix alba from a constructed wetland (Villette et al. 2019). Within that work, several TEL metabolites (based on hydroxylation, oxidation, and demethylation) as well as their distribution within Salix alba leaves using mass spectrometry imaging were described.
In the present paper, we conducted experiments using several plants grown either directly in water or hydroponically. Thereby the potential metabolites were characterized by their chromatographic behavior (retention time in reversed-phase high-performance liquid chromatography (RP-HPLC)), their accurate mass, and fragmentation in MS/MS experiments. In addition collision cross sections (DTCCSN2) using drift-tube ion-mobility quadrupole time-of-flight mass spectrometry (DTIM-QTOF-MS) with nitrogen as drift gas were measured. Finally, garden cress (Lepidium sativum) was selected as a model for research directed towards setting up an analytical method allowing the investigation of TEL uptake and metabolism in plants under environmentally relevant conditions. To evaluate the proposed methodology, cress plants were cultivated in water sampled from the receiving water course of a local WWTP.
Experimental
Chemicals and materials
Telmisartan (TEL) 80 mg (for structure see Fig. 1) was obtained as pharmaceutical preparation from Ratiopharm (Ulm, Germany). One tablet containing 80 mg TEL was homogenized with mortar and pestle. Sixty milligrams of the material (corresponding to 10 mg TEL) was used to prepare a 1000 mg L−1 stock solution in methanol. Further solutions of 100 mg L−1 and 10 mg L−1 were prepared by diluting the stock solution with Milli-Q water. For plant treatment, the solutions were further diluted with tap water. TEL-d3 (internal standard) was supplied by Cayman Chemical Company (Michigan, USA); from this, a 10 mg L−1 stock solution was prepared in methanol.
Methanol and acetonitrile were delivered by VWR (Vienna, Austria). Formic acid (eluent additive for LC-MS, 98%) was purchased from Sigma-Aldrich (Steinheim, Germany). Purified water was produced by a Milli-Q water purification system (Millipore, Bedford, MA, USA).
Plant cultivation and treatment
The cress seeds (Lepidium sativum L.) were supplied by Sperli (Everswinkel, Germany), cyprus grass (Cyperus zumula) was from OBI (Linz, Austria), pea (Pisum sativum, cv. Premium) from MoravoSeed CZ Corporation (Czech Republic), and maize (Zea mays, cv. Agnan) from Oseva Agro Brno Ltd. (Czech Republic). For detection of TEL-related metabolites, samples were cultivated under hydroponic conditions in 10 mg L−1 TEL solution and harvested after 7–12 days of cultivation. A more detailed description of the cultivation procedure can be found in a previously published report (Mlynek et al. 2021). Water plants, namely three-part pennywort (Hydrocotyle cf. tripartite) and parrot leaf (Alternanthera reineckii) were cultivated in 30 cm × 20 cm × 20 cm aquaria filled with tap water. For treatment, a certain amount of the plant was transferred to an Erlenmeyer flask containing in 10 mg L−1 TEL in tap water.
Cress was in addition cultivated in solutions of 10 μg L−1, 1 μg L−1, 100 ng L−1, and 10 ng L−1 TEL solutions and water from a WWTP effluent. For germination, seeds were soaked overnight in the corresponding growing solution and afterwards placed on cress-growing containers and humidified once a day for 2 days. During the germination period, the containers were kept in small greenhouses placed on the lab-window bench. Afterwards, the top of the greenhouse was removed in order to ensure proper air circulation. The containers were filled with the growing medium, which was replenished approximately every 2 days. This consumed in total 0.6 l for each dish.
Preparation of plant extracts for tentative identification of metabolites
The plants were harvested after 7–12 days in total and for cress, pea and maize roots and upper part were separated. The plant material was cleaned three times with tap water and dried with paper tissue. 1.5 g (wet weight) of the plant material was transferred into a 15-mL centrifugation tube. Subsequently, 3 mL of the extraction solvent as well as three steel beads with a diameter of 5 mm was added. As extraction solvent acetonitrile/water (v/v 1/2) was used. Samples were homogenized using a ball mill (“Star Beater,” VWR, Vienna Austria) for 15 min at 20 Hz. The homogenized samples were centrifuged for 20 min at 4000 rpm. The supernatant was attained with a syringe and subsequently filtered into 1.5-mL HPLC glass vials using a 0.45-μm syringe filter. The samples were stored at −80 °C until analysis.
Internal standard
TEL-d3 was implemented as internal standard (ISTD) for the targeted analysis. The 1000 ng L−1 ISTD was added to the wastewater as well as to the extracts of cress cultivated in wastewater (in the ratio ISTD/sample 1/10, v/v) directly before analysis.
Analysis of WWTP effluent samples
0.5 L of water was filtered through a 0.45-μm filter and passed through an Oasis HLB (Waters, Milford USA) solid-phase extraction cartridge. The extract was eluted using 5 mL of MeOH which was subsequently evaporated to (almost) dryness. After re-dissolving the residue in 500 μL of purified water, the sample was analyzed using the HPLC/DTIM-QTOF-MS instrument described below.
HPLC/DTIM-QTOF-MS and HPLC/QqQ-MS/MS instrumentation
The samples were separated by RP-HPLC using an Agilent 1260 HPLC system from Agilent Technologies (Waldbronn, Germany) equipped with a degasser, a quaternary pump, and an autosampler. The separation was performed on a Poroshell 120 EC-C18 column (3 × 150 mm, particle size 2.7 μm, Agilent) that was protected with a C18 guard column (4 × 3 mm, particle size 3 μm) from Phenomenex (Aschaffenburg, Germany).
For HPLC separation, a water/methanol gradient was applied. Starting conditions were set to 95% solvent A (water with 0.1% formic acid) and 5% solvent B (methanol). From minute 0 to 15, solvent B was increased to 100% and kept constant for 5 min. The column was re-equilibrated for 5 min, resulting in a total run time of 25 min. The flow rate was set to 0.4 mL min−1, the temperature of the column heater was 30 °C, and an injection volume of 20 μL was used.
For the tentative identity confirmation of metabolites, the HPLC system was hyphenated with an Agilent 6560 DTIM-QTOF LC-MS/MS (operated either in the “QTOF only” or ion-mobility MS mode) equipped with a Dual AJS ESI source (Agilent Technologies, Waldbronn, Germany).
The DTIM-QTOF-MS was tuned in the “fragile ion” mode and operated in the positive ionization mode with the following source parameters: drying gas temperature 300 °C, drying gas flow rate 10 L min−1, nebulizer pressure 50 psi, sheath gas temperature 300 °C, sheath gas flow rate 10 L min−1, capillary voltage 3500 V, nozzle voltage 1000 V, and fragmentor 425 V. For MS/MS experiments, nitrogen was used as collision gas and collision energies of 20 V and 30 V were applied.
The DTCCSN2 values were obtained using nitrogen as drift gas and the following parameters for the DTIM device: 4-bit multiplexing, frame rate 0.9 frames s−1, IM transient rate 18 transients frame−1, max drift time 60 ms, trap fill time 3900 μs, and trap release time 250 μs. Calibration was performed according to a “single-field” approach, allowing the determination of the DTCCSN2 values. In order to relate the measured drift times to known and standardized DTCCSN2 values of the calibrant analytes, a tune mix calibrant was measured (applying the same conditions as for the samples) before analyzing the actual sample. Drift tube parameters (for “single-field” measurements) were the following: drift tube entrance 1567 V, drift tube exit 217 V, rear funnel entrance 210.5 V, and rear funnel exit 38 V.
For the targeted analysis, an HPLC was coupled to an Agilent 6460 triple quadrupole QqQ-MS/MS (Agilent Technologies, Waldbronn, Germany) equipped with an AJS ESI source.
The QqQ MS/MS system was operated in the positive ionization mode. Applied parameters were as follows: capillary voltage 4000 V, drying gas flow rate 10 L min−1, drying gas temperature 300 °C, nebulizer pressure 45 psi. The optimized parameters (selected transitions, fragmentor voltages and collision energies) for the multiple reaction monitoring (MRM) mode are given in Table 1.
Data processing
For data evaluation, Agilent MassHunter Qualitative Analysis B.07.00, MassHunter Quantitative Analysis B.10.1, MassHunter PCDL Manager B.08.00, PNNL PreProcessor (2020.03.23), and the IM-MS Browser B.10.00 were used.
A database (PCDL) was created, similar as described in a previous study (Mlynek et al. 2021). Therefore, TEL or hydroxylated TEL were in silico combined with common building blocks of phase II metabolites like glucose or malonic acid in every combination and any number possible. This database is used for screening the MS spectra of treated plant samples. The results were verified by a targeted MS/MS looking at the accurate masses and fragmentation pattern. With the fragmentation pattern, possible sum formulas for metabolites were reconstructed from mass losses.
Ion-mobility (IM) data files were first demultiplexed using the PNNL PreProcessor software. Afterwards, the data were calibrated with the recorded single field tune using IM-MS browser. The drift times and the DTCCSN2 were determined by first performing a feature extraction (Find features IMFE). Parameters were processing chromatographic, isotope model common organic molecules, limit charge state z ≤ 1, ion intensity ≥ 1. The drift times and the collision cross sections of all features were automatically determined by the software. Within all the features found by the software, the analytes of interest were identified according to their m/z values and their retention times.
Results and discussion
Tentative identification of TEL metabolites by DTIM-QTOF-MS
Extracts from plants cultivated in the presence of 10 mg L−1 TEL, either hydroponically (pea, cress, maize) or directly in water (pennywort, parrot leaf), were screened for the presence of potential TEL-related metabolites using HPLC-DTIM-QTOF-MS. Employing the lab-made database, altogether nine TEL-derived substances were found in addition to the parent drug, whereby positive hits were further confirmed by MS/MS measurements. From published work, it was known that upon uptake by plants TEL can be hydroxylated (either one or two times), demethylated, or decarboxylated (Villette et al. 2019). Based on these findings, we could propose suggestions for tentative sum formulas for seven hits — whereas in two cases (i.e., m/z 777.3107 and m/z 793.3096), no proposal for a tentative structure could be made. The tentatively identified compounds comprise phase I + II metabolites resulting from the addition of glucose or malonic acid (or a combination of both) either to TEL (most probably to the carboxylate group) or to hydroxylated TEL (TEL-OH), where the hydroxyl group is the most probable site for conjugation. An overview of metabolites detected in these plants is given in Table 2.
For further studies, garden cress was selected as the best suited model plant. As the basic concept of the present study was to allow the analysis of plants grown under environmentally relevant conditions besides a more in-depth investigation on the presence of TEL-related substances in the extract, also a MRM method was developed on a QqQ-MS/MS instrument. Relevant information on the outcome of these investigations is summarized in Table 1. Regarding relative intensities (due to the lack of standard substances, only signal areas could be compared) of metabolite signals, TEL-glucose (TEL-glc) could be identified as the major metabolite, showing an abundance between 80 and 100% compared to that of TEL. The peak area for TEL combined with glucose and malonic acid (mal) was between 14 and 20% of that obtained for the parent drug. For the other compounds, substantially smaller peaks with areas of 1% and below (compared to TEL) were registered.
For an overview of retention time ranges, accurate mass (of the protonated species), mass error, and DTCCSN2, see Table 1. In this table, no exact retention times but retention time ranges are provided as for several compounds more than one signal with identical accurate mass and MS/MS spectrum but slightly different retention time and also slightly different DTCCSN2 was detected. This was in accordance with previous observations and can be explained by the presence of isomers showing variations in the three-dimensional structure of the molecules, thereby influencing both the chromatographic behavior and the drift times in the ion-mobility device (Mlynek et al. 2020). An exemplary MS/MS spectrum for TEL-glc-mal, a metabolite detected in the cress extracts can be seen in Fig. 2. For the two unidentified metabolites, accurate masses of m/z 777.3107 and m/z 793.3096 were determined. Using the PCDL, the following suggestions were obtained: TEL with glucose and malonic acid with an additional methyl group or succinic instead of malonic acid for the first mass, and the same composition but originating from TEL-OH instead of TEL for the second. The ∆ m/z of 15.9989 suggesting the presence of an additional oxygen in the second molecule can be seen as a further hint strengthening this proposal. Unfortunately the latter assumption is not bolstered by the MS/MS data as in both cases, employing collision energies of 20 V, a direct transition to TEL (and not to TEL-OH) with subsequent loss of a water molecule was observed (see Fig. 3). These data strongly suggested that both tentative metabolites originated from TEL and the additional oxygen was part of the moiety attached to a TEL molecule. A comparison of the MS/MS spectra obtained for the two unknown metabolites is depicted in Fig. 3.
Targeted analysis of the parent drugs and their metabolites by implementing an HPLC-QqQ-MS/MS method
Based on the results described in the “Tentative identification of TEL metabolites by DTIM-QTOF-MS” section, investigations on the trace level detection of TEL and its tentative metabolites in extracts from cress plants were performed. The main goal of these experiments was to find out whether the developed approach might be used for plantlets grown in actual reclaimed waters or waters drawn from the effluents of WWTPs. Therefore, several batches of cress plants were cultivated in parallel hydroponically using water containing TEL at levels between 10 μg L−1 (3 orders of magnitude less than the concentration level employed for the search for potential metabolites) and 10 ng L−1. Thereby the parent drug and TEL-glc were found in all plants whereas TEL-OH-glc was found in the plants cultivated in the10 μg L−1 and 1 μg L−1 solution only. TEL-OH could exclusively be detected in the cress from the 10 μg L−1 growing medium. To evaluate parameters relevant for quantitative analysis of TEL, two calibration curves (one in water and one in cress extract matrix) were constructed between 20 and 200 ng L−1. Both showed a correlation coefficient of 0.99 or better. The fact that a slope of around 16.200 was achieved for the standards in water compared to around 11.700 for the matrix matched calibration suggested the presence of ionization suppression in the ESI source caused by plant matrix compounds. LODs for TEL in water and cress matrix were both found to be 9 ng L−1. In order to further evaluate the analytical procedure, five sets of plants were cultivated, harvested, and spiked with TEL and plant extracts were prepared on three consecutive days. These extracts were subsequently analyzed three times each by HPLC-QqQ-MS/MS. These experiments led to an average recovery of 69% with a relative standard deviation of (RSD) 8.7% on day one, 68% with a RSD of 8.6% on day two, and 70% with a RSD of 8.8% on day three for TEL.
Analysis of garden cress cultivated in water from a WWTP effluent
A water sample was drawn from the effluent of a WWTP and analyzed qualitatively (and also quantitatively for TEL) using HPLC-QTOF-MS after appropriate sample pre-treatment (for exact procedure see the “HPLC/DTIM-QTOF-MS and HPLC/QqQ-MS/MS instrumentation” section). An overview of the pharmaceuticals detected in this water sample (employing a semi-targeted approach) is provided in Table S1. HPLC-QqQ-MS analysis revealed a TEL content of 90 ng L−1 in the effluent sample that was further employed for growing the cress plantlets hydroponically. The water sample was also checked for the presence of TEL-related metabolites (in particular TEL-OH) but none of them could be detected. Subsequently, cress plantlets were grown in the water from the WWTP effluent and harvested, and in a first step extracts from the plants were screened employing the RP-HPLC MRM QqQ MS method already used for the experiments described in the “Targeted analysis of the parent drugs and their metabolites by implementing an HPLC-QqQ-MS/MS method” section. Thereby, as was expected, only TEL and TEL-glc could be detected. All other metabolites were below or very close to the LOD, leading to m/z traces that could not be quantified properly. The TEL concentration determined in three independently grown batches of cress, employing a matrix matched calibration curve, was 390 pg g−1 for cress roots (wet weight) with a relative standard deviation (RSD) of 25%. In this context, it must be taken into account that apart from the analytical methodology employed, also biological variations contributed to this RSD. For comparison 90 μL of the cress extract was spiked with 10 μL of a 1000 ng L−1 TEL-d3 standard solution to perform quantitative analysis also using the standard addition method. Evaluation of these experiments led to a TEL concentration of 550 pg per g cress with an RSD of 24%. A different situation was encountered for TEL-glc as no standard for quantitation was available. Here only peak areas could be compared. The fact that for the plants grown in a 100 ng L−1 solution of TEL (a concentration very close to the one in the WWTP effluent), a 50% higher peak area was registered for TEL-glc than for those grown in water from a WWTP effluent might be explained by the presence of other xenobiotics in the latter, partially utilizing the same biological pathways required for glycosylation of the parent substance.
Conclusions
In the present work, we managed to tentatively identify eight TEL-related metabolites formed in three different plants cultivated hydroponically and in two water plants. In both cases, growing media containing TEL in the mg L−1 range were used. Based on these findings, further investigations were conducted, using garden cress as a model plant, focusing on the possibility to transfer this knowledge also to environmentally relevant samples commonly in contact with emerging contaminants like TEL in much lower concentrations. The approach followed was to cultivate garden cress hydroponically in a water sample drawn from a WWTP effluent. In the corresponding samples, TEL as well as one major metabolite (TEL-glc) could be identified and TEL quantified using a matrix matched calibration curve as well as the standard addition method employing a deuterated TEL standard. As WWTP effluent samples commonly contain a plethora of emerging pollutants in concentrations up to the high ng L−1 range, further work will be directed towards synergetic effects with respect to uptake and metabolization of widely used drugs in such plant models.
Data availability
All data and materials are included in this published article.
References
Alcalde-Sanz L, Gawlik BM (2017) Minimum quality requirements for water reuse in agricultural irrigation and aquifer recharge - towards a legal instrument on water reuse at EU level, Publications Office of the European Union 1–63
Bayer A, Asner R, Schüssler W, Kopf W, Weiß K, Sengl M, Letzel M (2014) Behavior of sartans (antihypertensive drugs) in wastewater treatment plants, their occurrence and risk for the aquatic environment. Environ Sci Pollut Res 21:10830–10839
Beltrán EM, Pablos MV, Fernández Torija C, Porcel MA, González-Doncel M (2020) Uptake of atenolol, carbamazepine and triclosan by crops irrigated with reclaimed water in a Mediterranean scenario. Ecotoxicol Environ Saf 191:110171
Buchberger WW (2011) Current approaches to trace analysis of pharmaceuticals and personal care products in the environment. J Chromatogr A 1218:603–618
Castro G, Rodríguez I, Ramil M, Cela R (2019) Selective determination of sartan drugs in environmental water samples by mixed-mode solid-phase extraction and liquid chromatography tandem mass spectrometry. Chemosphere 224:562–571
Cotton J, Leroux F, Broudin S, Poirel M, Corman B, Junot C, Ducruix C (2016) Development and validation of a multi residue method for the analysis of more than 500 pesticides and drugs in water based on on-line and liquid chromatography coupled to high resolution mass spectrometry. Water Res 104:20–27
European Comission, European Parliament (2008), DIRECTIVE 2008/105/EC. https://eur-lex.europa.eu/eli/dir/2008/105/oj.
European Comission, European Parliament (2013), DIRECTIVE 2013/39/EC. http://data.europa.eu/eli/dir/2013/39/oj.
Emhofer L, Himmelsbach M, Buchberger W, Klampfl CW (2018) Insights into the uptake, metabolization and translocation of four non-steroidal anti-inflammatory drugs in cress (Lepidium sativum) by HPLC-MS2. Electrophoresis 39:1294–1300
Emhofer L, Himmelsbach M, Buchberger W, Klampfl CW (2019) HPLC drift-tube ion-mobility quadrupole time-of-flight / mass spectrometry for the identification and characterization of metabolites from three statins (lipid lowering drugs) in the mode plant garden cress (Lepidium sativum). J Chromatogr A 1592:122–132
Fu Q, Malchi T, Carter LJ, Li H, Gan J, Chefetz B (2019) Pharmaceutical and personal care products: from wastewater treatment into agro-food systems. Environ Sci Technol 53:14083–14090
Giebułtowicz J, Stankiewicz A, Wroczyński P, Nałęcz-Jawecki G (2016) Occurrence of cardiovascular drugs in the sewage-impacted Vistula River and in tap water in the Warsaw region (Poland). Environ Sci Pollut Res 23:24337–24349
Hignite C, Azarnoff DL (1977) Drugs and drug metabolites as environmental contaminants - chlorophenoxyisobutyrate and salicylic-acid in sewage water effluent. Life Sci 20:337–341
Keerthanan S, Jayasinghe C, Kumar Biswas J, Vithanage M (2021) Pharmaceutical and personal care products (PPCPs) in the environment: plant uptake, translocation, bio-accumulation, and human health risks. Crit Rev Environ Sci Technol:1–38. https://doi.org/10.1080/10643389.2020.1753634
Kinney CA, van den Heuvel B (2020) Translocation of pharmaceuticals and personal care products after land application of biosolids. Curr Opin Environ Sci Health 14:23–30
Klampfl CW (2019) Metabolization of pharmaceuticals by plants after uptake from water and soil: a review, Trends Anal. Chem. 111:13–26
Klampfl CW, Buchberger W, Mlynek F, Himmelsbach M (2021) Metabolization of pharmaceuticals by plants after uptake from water. In: Solsona SP, Montemurro N, Chiron S, Barceló D (eds) Interaction and fate of pharmaceuticals in soil-crop systems: the impact of reclaimed wastewater, Hdb Env Chem. https://doi.org/10.1007/698_2020_629Springer-Nature
Lindberg RH, Östman M, Olofsson U, Grabic R, Fick J (2014) Occurrence and behaviour of 105 active pharmaceutical ingredients in sewage waters of a municipal sewer collection system. Water Res 58:221–229
Mlynek F, Himmelsbach M, Buchberger W, Klampfl CW (2020) A new analytical workflow using HPLC with drift-tube ion-mobility quadrupole time-of-flight/mass spectrometry for the identification of drug-related metabolites in plants. Anal Bioanal Chem 412:1817–1824
Mlynek F, Himmelsbach M, Buchberger W, Klampfl CW (2021) A fast-screening approach for the tentative identification of drug-related metabolites from three non-steroidal anti-inflammatory drugs in hydroponically grown edible plants by HPLC-drift-tube-ion-mobility quadrupole time-of-flight mass spectrometry. Electrophoresis 42:482–489
Montemurro N, Postigo C, Lonigro A, Perez S, Barceló D (2017) Development and validation of an analytical method based on liquid chromatography–tandem mass spectrometry detection for the simultaneous determination of 13 relevant wastewater-derived contaminants in lettuce. Anal Bioanal Chem 409:5375–5387
Nuel M, Laurent J, Bois P, Heintz D, Wanko A (2018) Seasonal and ageing effect on the behavior of 86 drugs in a full-scale surface treatment wetland: removal efficiencies and distribution in plants and sediments. Sci Total Environ 615:1099–1109
Petrie B, McAdam EJ, Scrimshaw MD, Lester JN, Cartmell E (2013) Fate of drugs during wastewater treatment, Trends Anal. Chem. 49:145–159
Pico Y, Alfarham A, Barcelo D (2017) Analysis of emerging contaminants and nanomaterials in plant materials following uptake from soils, Trends Anal. Chem. 94:173–189
Puckowski K, Mioduszewska K, Łukaszewicz P, Borecka M, Caban M, Maszkowska J, Stepnowski P (2016) Bioaccumulation and analytics of pharmaceutical residues in the environment: a review. J Pharm Biomed Anal 127:232–255
Reddy Pullagurala VL, Rawat S, Adisa IO, Hernandez-Viezcas JA, Peralta-Videa JR, Gardea-Torresdey JL (2018) Plant uptake and translocation of contaminants of emerging concern in soil. Sci Total Environ 636:1585–1596
Richardson SD, Ternes TA (2014) Water analysis: Emerging contaminants and current issues. Anal Chem 86:2813–2848
Riemenschneider C, Seiwert B, Moeder M, Schwarz D, Reemtsma T (2017) Extensive transformation of the pharmaceutical carbamazepine following uptake into intact tomato plants. Environ Sci Technol 51:6100–6109
Rzymski P, Drewek A, Klimaszyk P (2017) Pharmaceutical pollution of the aquatic environment: an emerging and enormous challenge. Limnol Rev 17:97–107
Villette C, Maurer L, Wanko A, Heintz D (2019) Xenobiotics metabolization in Salix alba leaves uncovered by mass spectrometry imaging. Metabolomics 15:122
Acknowledgements
Part of this work was supported by the Austrian Science Fund (FWF) project I-3046 (Pharmaceuticals in the Environment and their Interaction with Plants). The authors thank Mrs. Laura Mairinger for the qualitative analysis of WWTP effluent water samples.
Funding
Open access funding provided by Johannes Kepler University Linz.
Author information
Authors and Affiliations
Contributions
TL has performed the experimental work and assisted in writing the manuscript; MH provided help in performing and interpretation of LC-MS/MS experiments; FM supervised the experimental work; WB and CWK designed the study and wrote up the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Ester Heath
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 12 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lang, T., Himmelsbach, M., Mlynek, F. et al. Uptake and bio-transformation of telmisartan by cress (Lepidium sativum) from sewage treatment plant effluents using high-performance liquid chromatography/drift-tube ion-mobility quadrupole time-of-flight mass spectrometry. Environ Sci Pollut Res 28, 50790–50798 (2021). https://doi.org/10.1007/s11356-021-14289-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-021-14289-4