Abstract
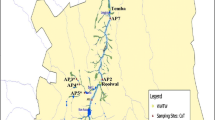
The identification of fecal pollution in aquatic ecosystems is one of the requirements to assess the possible risks to human health. In this report, physicochemical parameters, Escherichia coli enumeration and Methanobrevibacter smithii nifH gene quantification were conducted at 13 marine waters in the coastal beaches of Rio de Janeiro, Brazil. The pH, turbidity, dissolved oxygen, temperature, and conductivity, carried out by mobile equipment, revealed varied levels due to specific conditions of the beaches. The bioindicators’ enumerations were done by defined substrate method, conventional, and real-time PCR. Six marine beach sites (46 %) presenting E. coli levels in compliance with Brazilian water quality guidelines (<2500 MPN/100 mL) showed nifH gene between 5.7 × 109 to 9.5 × 1011 copies. L−1 revealing poor correlation between the two approaches. To our knowledge, this is the first inquiry in qPCR using nifH gene as a biomarker of human-specific sources of sewage pollution in marine waters in Brazil. In addition, our data suggests that alternative indicator nifH gene could be used, in combination with other markers, for source tracking studies to measure the quality of marine ecosystems thereby contributing to improved microbial risk assessment.

Similar content being viewed by others
References
Ahmed W, Sidhu JPS, Toze S (2012) Evaluation of the nifH gene marker of Methanobrevibacter smithii for the detection of sewage pollution in environmental waters in Southeast Queensland, Australia. Environmental science & technology 46(1):543–550
American Public Health Association–APHA; American Water Works Association–AWWA; Water Environment Federation–WEF (2012). Standard methods for the examination of water and wastewater. 22th ed. Washington DC.
Ballesté E, Bonjoch X, Belanche LA, Blanch AR (2010) Molecular indicators used in the development of predictive models for microbial source tracking. Appl Environ Microbiol 76(6):1789–1795
Bernhard AE, Goyard T, Simonich MT, Field KG (2003) Application of a rapid method for identifying fecal pollution sources in a multi-use estuary. Water Res 37:909–913
Bianco K, Barreto C, Oliveira SS, Pinto LH, Albano RM, Miranda CC, Clementino MM (2015) Fecal pollution source tracking in waters intended for human supply based on archaeal and bacterial genectic markers. J Water Health 13(4):985–995
Boehm AB, Ashbolt NJ, Colford JM, Dunbar LE, Fleming LE, Gold MA, Hansel JA, Hunter PR et al (2009) A sea change ahead for recreational water quality criteria. J Water Health 7:9–20
Clementino MM, Fernandes CC, Vieira RP, Cardoso AM, Polycarpo CR, Martins OB (2007) Archaeal diversity in naturally occurring and impacted environments from a tropical region. J Appl Microbiol 103(1):141–151
Cole D, Long SC, Sobsey MD (2003) Evaluation of Fþ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl Environ Microbiol 69(11):6507–6514
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlesfsen L, Sargent M, Gill SR, Nelson KE et al (2005) Diversity of the human intestinal microbial flora. Science 308:1635–1638
Fewtrell L, Kay D (2015) Recreational water and infection: a review of recent findings. Current Environmental Health Reports 2(1):85–94
Field KG, Samadpour M (2007) Fecal source tracking, the indicator paradigm, and managing water quality. Water Res 41(16):3517–3538
Foley SL, Lynne AM, Nayak R (2009) Molecular typing methodologies for microbial source tracking and epidemiological investigations of Gram-negative bacterial foodborne pathogens. Infect Genet Evol 9(4):430–440
Fong TT, Griffin DW, Lipp EK (2005) Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Appl Environ Microbiol 71(4):2070–2078
Gourmelon M, Caprais MP, Ségura R, Le Mennec C, Lozach S, Piriou JY, Rincé A (2007) Evaluation of two library-independent microbial source tracking methods to identify sources of fecal contamination in French estuaries. Appl Environ Microbiol 73(15):4857–4866
Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A (2014) Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38(1):1–40
Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP (2005) Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase change reaction and membrane filter culture analysis. Water Res 39:559–568
Jannasch HW (1968) Competitive elimination of Enterobacteriaceae from seawater. Appl Microbiol 16:1616–1618
Johnston C, Ufnar JA, Griffith JF, Gooch JA, Stewart JR (2010) A real-time qPCR assay for the detection of the nifH gene of Methanobrevibacter smithii, a potential indicator of sewage pollution. J Appl Microbiol 109(6):1946–1956
Lai MC, Lin CC, Yu PH, Huang YF, Chen SC (2004) Methanocalculus chunghsingenisis sp. Nov., isolated from an estuary and a marine fishpond in Taiwan. Int J Syst Evol Microbiol 54:183–118
Layton BA, Cao Y, Ebentier DL, Hanley K, Ballesté E, Brandão J, Byappanahalli M, Converse R, Farnleitner AH, Gentry-Shields J, Gidley ML, Gourmelon M, Lee CS, Lee J, Lozach S, Madi T, Meijer WG, Noble R, Peed L, Reischer GH, Rodrigues R, Rose JB, Schriewer A, Sinigalliano C, Srinivasan S, Stewart J, Werfhorst LC, Wang D, Whitman R, Wuertz S, Jay J, Holden PA, Boehm AB, Shanks O, Griffith JF (2013) Performance of human fecal anaerobe-associated PCR-based assays in a multi-laboratory method evaluation study. Water Res 47(Issue 18):6897–6908
Lebaron P, Cournoyer B, Lemarchand K, Nazaret S, Servais P (2015) Environmental and human pathogenic microorganisms. In: Environmental microbiology: fundamentals and applications. Springer, Netherlands, pp. 619–658
McQuaig SM, Scott TM, Lukasik JO, Paul JH, Harwood VJ (2009) Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl Environ Microbiol 75:3379–3388
Ogram A, Sayler GS, Barkay T (1987) The extraction and purification of microbial DNA from sediments. J Microbiol Methods 7:57–66
Oliveira BSSD, Cunha ACD (2014) Correlation between water quality and precipitation variability in the southern state of Amapá. Rev Ambient Água 9(2):261–275
Rasmussen R (2001) In: Meuer S, Wittwer C, Nakagawara K (eds) Quantification on the LightCycler. Springer-Verlag, Berlin, pp. 21–34
Rozen Y, Belkin S (2001) Survival of enteric bacteria in seawater. FEMS Microbiol Rev 25(5):513–529
Santo Domingo JW, Bambic DG, Edge TA, Wuertz S (2007) Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution. Wat Res 41:3539–3552
Savichtcheva O, Okabe S (2006) Alternative indicators of fecal pollution: relations with pathogens and conventional indicators, current methodologies for direct pathogen monitoring and future application perspectives. Water Res 40:2463–2476
Schmiegelow, J. M. M. (2004). The Blue Planet: An Introduction Marine Science. Brazil.
Scott TM, Rose JB, Jenkins TM, Farrah SR, Luhasik J (2002) Microbial source tracking: current methodology and future directions. Appl Environ Microbiol 68:5796–5803
Scott TM, Jenkins TM, Lukasik J, Rose JB (2005) Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ Sci Technol 1(39):283–287
Shanks OC, Nietch C, Simonich M, Younger M, Reynolds D, Field KG (2006) Basin-wide analysis of the dynamics of fecal contamination and fecal source identification in fecal pollution source tracking in waters intended for human supply journal of water and health in Tillamook Bay, Oregon. Appl Environ Microbiol 72(8):5537–5546
Smalla K, Cresswell N, Mendonca-Hagler LC, Wolters A, van Elsas JD (1993) Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J Appl Bacteriol 74:78–85
Stewart-Pullaro J, Daugomah JW, Chestnut DE, Graves DA, Sobsey MD, Scott GI (2006) F+ RNA coliphage typing for microbial source tracking in surface waters. J Appl Microbiol 101(5):1015–1026
Stoeckel DM, Harwood VJ (2007) Performance, design, and analysis in microbial source tracking studies. Appl Environ Microbiol 73(8):2405–2415
Ufnar JA, Wang SY, Christiansen JM, Yampara-Iquise H, Carson CA, Ellender RD (2006) Detection of the nifH gene of Methanobrevibacter smithii: a potential tool to identify sewage pollution in recreational waters. J Appl Microbiol 101(1):44–52
Walters SP, Yamahara KM, Boehm AB (2009) Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms. Water Res 43:4929–4939
Whitman RL, Ge Z, Nevers MB, Boehm AB, Chern EC, Haugland RA, White EM (2010) Relationship and variation of qPCR and culturable enterococci estimates in ambient surface waters are predictable. Environ Sci Technol 44:5049–5054
Wimalawansa SA, Wimalawansa SJ (2014) Impact of changing agricultural practices on human health: chronic kidney disease of multi-factorial origin in Sri Lanka. Wudpecker J Agric Res 3(5):110–124
Acknowledgments
We gratefully acknowledge the Genome Sequencing Core-PDTIS/FIOCRUZ. This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Ensino Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), and INCQS/FIOCRUZ.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Rights and permissions
About this article
Cite this article
Oliveira, S.S., Sorgine, M.H.F., Bianco, K. et al. Detection of human fecal contamination by nifH gene quantification of marine waters in the coastal beaches of Rio de Janeiro, Brazil. Environ Sci Pollut Res 23, 25210–25217 (2016). https://doi.org/10.1007/s11356-016-7737-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-016-7737-3