Abstract
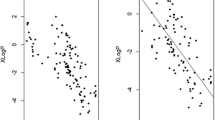
Metabolic profiling focuses on the analysis of a wide range of small endogenous molecules in order to understand the response of a living system to perturbations. Ultra high performance liquid chromatography–mass spectrometry is a widely employed profiling tool, but its application is limited by difficulties in identification of detected metabolites. Herein, we demonstrate how the prediction of retention time can help resolve this major issue. We describe a general approach that enables the generation of reliable quantitative structure retention relationship models tailored to specific chromatographic protocols. This methodology, applied to 442 experimentally characterised standards, employs a combination of random forest and support vector regression models with molecular interaction descriptors. In this unusual application, the Volsurf + molecular descriptors demonstrated a high ability to describe chromatographic retention. On external validation sets, and for a wide range of chemical classes, predicted values were in average within 13 % of the experimentally observed retention time. More importantly, the presented procedure reduced by more than 80 % the number of false putative identification, greatly improving metabolite identification. Furthermore, in 95 % of cases, the correct identification was promoted within the top three metabolite suggestions. This retention time prediction framework can be replicated by different laboratories to suit their profiling platforms and enhance the value of standard library by providing a new tool for compound identification.




Similar content being viewed by others
References
Almeida, T. M. G., Leitão, A., Montanari, M. L. C., & Montanari, C. A. (2005). The molecular retention mechanism in reversed-phase liquid chromatography of meso-ionic compounds by quantitative structure-retention relationships (QSRR). Chemistry & Biodiversity, 2(12), 1691–1700.
Baczek, T., & Kaliszan, R. (2009). Predictions of peptides’ retention times in reversed-phase liquid chromatography as a new supportive tool to improve protein identification in proteomics. Proteomics, 9(4), 835–847.
Beckonert, O., Keun, H. C., Ebbels, T. M. D., Bundy, J., Holmes, E., Lindon, J. C., & Nicholson, J. K. (2007). Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nature Protocols, 2(11), 2692–2703.
Berthold, M. R., Cebron, N., Dill, F., Gabriel, T. R., Kötter, T., Meinl, T., et al. (2009). KNIME—the Konstanz information miner: Version 2.0 and beyond. SIGKDD Explorations, 11(1), 26–31.
Boswell, P. G., Schellenberg, J. R., Carr, P. W., Cohen, J. D., & Hegeman, A. D. (2011a). Easy and accurate high-performance liquid chromatography retention prediction with different gradients, flow rates, and instruments by back-calculation of gradient and flow rate profiles. Journal of Chromatography A, 1218(38), 6742–6749.
Boswell, P. G., Schellenberg, J. R., Carr, P. W., Cohen, J. D., & Hegeman, A. D. (2011b). A study on retention “projection” as a supplementary means for compound identification by liquid chromatography-mass spectrometry capable of predicting retention with different gradients, flow rates, and instruments. Journal of Chromatography A, 1218(38), 6732–6741.
Breiman, L. (2001). Random Forests. Machine Learning, 45(1), 5–32.
Chang, C., & Lin, C. (2011). LIBSVM: A library for support vector machines. ACM Transactions on Intelligent Systems and Technology, 2(3), 1–27.
Creek, D. J., Jankevics, A., Breitling, R., Watson, D. G., Barrett, M. P., & Burgess, K. E. V. (2011). Toward global metabolomics analysis with hydrophilic interaction liquid chromatography–mass spectroscopy: Improved metabolite identification by retention time prediction. Analytical Chemistry, 83, 8703–8710.
Cruciani, G., Mannhold, R., Berellini, G., Carosati, E., & Benedetti, P. (2006). Chapter 8. Use of MIF-based VolSurf descriptors in physicochemical and pharmacokinetic studies. In G. Cruciani (Ed.), Molecular interaction fields: Applications in drug discovery and ADME prediction (pp. 171–196). Weinheim: Wiley.
De Vos, R. C. H., Moco, S., Lommen, A., Keurentjes, J. J. B., Bino, R. J., & Hall, R. D. (2007). Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nature Protocols, 2(4), 778–791.
Dimitrov, S., Dimitrova, G., Pavlov, T., Dimitrova, N., Patlewicz, G., Niemela, J., & Mekenyan, O. (2005). A stepwise approach for defining the applicability domain of SAR and QSAR models. Journal of Chemical Information and Modeling, 45(4), 839–849.
Dunn, W. B., Broadhurst, D. I., Atherton, H. J., Goodacre, R., & Griffin, J. L. (2011). Systems level studies of mammalian metabolomes: The roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chemical Society Reviews, 40(1), 387–426.
Ermondi, G., & Caron, G. (2012). Molecular interaction fields based descriptors to interpret and compare chromatographic indexes. Journal of Chromatography A, 1252, 84–89.
Fiehn, O. (2002). Metabolomics—the link between genotypes and phenotypes. Plant Molecular Biology, 48(1–2), 155–171.
Ghasemi, J., & Saaidpour, S. (2009). QSRR prediction of the chromatographic retention behavior of painkiller drugs. Journal of Chromatographic Science, 47(2), 156–163.
Golbraikh, A., Shen, M., Xiao, Z., Xiao, Y.-D., Lee, K.-H., & Tropsha, A. (2003). Rational selection of training and test sets for the development of validated QSAR models. Journal of Computer-Aided Molecular Design, 17(2–4), 241–253.
Golbraikh, A., & Tropsha, A. (2002). Beware of q2! Journal of Molecular Graphics and Modelling, 20(4), 269–276.
Gramatica, P., Cassani, S., Roy, P. P., Kovarich, S., Yap, C. W., & Papa, E. (2012). QSAR modeling is not “push a button and find a correlation”: A case study of toxicity of (benzo-)triazoles on algae. Molecular Information, 31(11–12), 817–835.
Hall, M., Frank, E., Holmes, G., Pfahringer, B., Reutemann, P., & Witten, I. H. (2009). The WEKA data mining software: An update. SIGKDD Exploration, 11(1), 10–18.
Héberger, K. (2007). Quantitative structure-(chromatographic) retention relationships. Journal of Chromatography A, 1158(1–2), 273–305.
Hu, R.-J., Liu, H.-X., Zhang, R.-S., Xue, C.-X., Yao, X.-J., Liu, M.-C., & Fan, B.-T. (2005). QSPR prediction of GC retention indices for nitrogen-containing polycyclic aromatic compounds from heuristically computed molecular descriptors. Talanta, 68(1), 31–39.
Jalali-Heravi, M., & Kyani, A. (2004). Use of computer-assisted methods for the modeling of the retention time of a variety of volatile organic compounds: a PCA-MLR-ANN approach. Journal of Chemical Information and Computer Sciences, 44(4), 1328–1335.
Kaliszan, R. (2007). QSRR: Quantitative structure-(chromatographic) retention relationships. Chemical Reviews, 107(7), 3212–3246.
Kind, T., & Fiehn, O. (2010). Advances in structure elucidation of small molecules using mass spectrometry. Bioanalytical Reviews, 2(1–4), 23–60.
Kohonen, T. (1990). The self-organizing map. Proceedings of the IEEE, 78(9), 1464–1480.
Lee, H. S., Kim, M. K., Lee, C., Kim, J., Choo, I. H., Woo, J. I., & Chong, Y. (2008). Chemometric studies on brain-uptake of PET agents via VolSurf analysis. Bulletin-Korean Chemical Society, 29(1), 61–68. doi:10.5012/bkcs.2008.29.1.061.
Luan, F., Xue, C., Zhang, R., Zhao, C., Liu, M., Hu, Z., & Fan, B. (2005). Prediction of retention time of a variety of volatile organic compounds based on the heuristic method and support vector machine. Analytica Chimica Acta, 537(1–2), 101–110.
Mihaleva, V. V., Verhoeven, H. A., de Vos, R. C. H., Hall, R. D., & van Ham, R. C. H. J. (2009). Automated procedure for candidate compound selection in GC-MS metabolomics based on prediction of Kovats retention index. Bioinformatics, 25(6), 787–794.
Nicholson, J. K., Lindon, J. C., & Holmes, E. (1999). “Metabonomics”: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica, 29(11), 1181–1189.
Nobeli, I., & Thornton, J. M. (2006). A bioinformatician’s view of the metabolome. BioEssays, 28(5), 534–545.
Nord, L. (1998). Prediction of liquid chromatographic retention times of steroids by three-dimensional structure descriptors and partial least squares modeling. Chemometrics and Intelligent Laborary Systems, 44(1–2), 257–269.
Perruccio, F., Mason, J. S., Sciabola, S., & Baroni, M. (2006). Chapter 4. FLAP: 4-Point pharmacophore fingerprints from GRID. In G. Cruciani (Ed.), Molecular interaction fields: Applications in drug discovery and ADME prediction (pp. 83–102). Weinheim: Wiley.
Put, R., & Vander Heyden, Y. (2007). Review on modelling aspects in reversed-phase liquid chromatographic quantitative structure-retention relationships. Analytica Chimica Acta, 602(2), 164–172.
Roberts, L. D., Souza, A. L., Gerszten, R. E., & Clish, C. B. (2012). Targeted metabolomics. In F. M. Ausubel (Ed), Current protocols in molecular biology (Chapter 30, Unit 30.2.1–24.)
Sahigara, F., Ballabio, D., Todeschini, R., & Consonni, V. (2013). Defining a novel k-nearest neighbours approach to assess the applicability domain of a QSAR model for reliable predictions. Journal of Cheminformatics, 5(1), 27–36.
Sahigara, F., Mansouri, K., Ballabio, D., Mauri, A., Consonni, V., & Todeschini, R. (2012). Comparison of different approaches to define the applicability domain of QSAR models. Molecules, 17(5), 4791–4810.
Shinoda, K., Sugimoto, M., Yachie, N., Sugiyama, N., Masuda, T., Robert, M., & Tomita, M. (2006). Prediction of liquid chromatographic retention times of peptides generated by protease digestion of the Escherichia coli proteome using artificial neural networks. Journal of Proteome Research, 5(12), 3312–3317.
Stein, S. E., Heller, S. R., & Tchekhovskoi, D. (2003). An open standard for chemical structure representation: The IUPAC chemical identifier. In Proceedings of the 2003 International Chemical Information Conference (Nimes), Infonortics (pp. 131–143).
Subirats, X., Rosés, M., & Bosch, E. (2007). On the effect of organic solvent composition on the pH of buffered HPLC mobile phases and the pKa of analytes—a review. Separation & Purification Reviews, 36(3), 231–255.
Sugimoto, M., Hirayama, A., Robert, M., Abe, S., Soga, T., & Tomita, M. (2010). Prediction of metabolite identity from accurate mass, migration time prediction and isotopic pattern information in CE-TOFMS data. Electrophoresis, 31(14), 2311–2318.
Tropsha, A. (2010). Best practices for QSAR model development, validation, and exploitation. Molecular Information, 29(6–7), 476–488.
Tropsha, A., Gramatica, P., & Gombar, V. K. (2003). The importance of being earnest: Validation is the absolute essential for successful application and interpretation of QSPR models. QSAR & Combinatorial Science, 22(1), 69–77.
Volsurf + 1.0.6 manual. Molecular Discovery, UK.
Want, E. J., Wilson, I. D., Gika, H., Theodoridis, G., Plumb, R. S., Shockcor, J., & Nicholson, J. K. (2010). Global metabolic profiling procedures for urine using UPLC-MS. Nature Protocals, 5(6), 1005–1018.
Wishart, D. S., Knox, C., Guo, A. C., Eisner, R., Young, N., Gautam, B., et al. (2009). HMDB: A knowledgebase for the human metabolome. Nucleic Acids Research, 37((Database issue)), D603–D610.
Worth, A., Bassan, A., Gallegos, A., Netzeva, T., Patlewicz, G., Pavan, M. et al. (2005). The characterisation of (Quantitative) Structure-Activity Relationships: Preliminary guidance. In ECB Report EUR 21866: European Commission, Joint Research Center (p. 95)
Zamora, I., Oprea, T., Cruciani, G., Pastor, M., & Ungell, A.-L. (2003). Surface descriptors for protein-ligand affinity prediction. Journal of Medicinal Chemistry, 46(1), 25–33.
Acknowledgments
The authors would like to thank Dr. Bernard Walther (Director Center of Excellence in PK) and Dr Claire Boursier-Neyret (Head of non-clinical PK) for their support during this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Human and animal informed consent
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
All authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wolfer, A.M., Lozano, S., Umbdenstock, T. et al. UPLC–MS retention time prediction: a machine learning approach to metabolite identification in untargeted profiling. Metabolomics 12, 8 (2016). https://doi.org/10.1007/s11306-015-0888-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-015-0888-2