Abstract
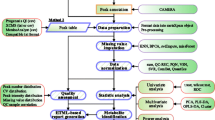
Multiple reaction monitoring (MRM)-based targeted metabolomics can simultaneously analyze up to hundreds of metabolites with high-throughput, good reproducibility, and wide dynamic range. However, when hundreds or thousands of MRM transitions are measured with tens to hundreds of biological samples, the complexity of MRM dataset acquired is no longer amenable to manual evaluation, and presents a challenge for targeted metabolomics. Here, we developed an R package, namely MRMAnalyzer, to process large set of MRM-based targeted metabolomics data automatically without any manual intervention. To demonstrate our MRMAnalyzer program, we first developed a targeted metabolomic method that simultaneously analyzes 182 metabolites in one 15-min LC run, and demonstrated the data processing procedures using MRMAnalyzer. The data processing steps include “pseudo” accurate m/z transformation, peak detection and alignment, metabolite identification, quality control check and statistical analysis. Finally, a targeted metabolomic assay was designed and integrated with MRMAnalyzer to profile the metabolic changes in Escherichia coli subjected to the protein expression. The generated MRM dataset consisting of more than 8000 MRM transitions were readily processed using MRMAnalyzer within 20 min without any manual intervention. Fourty seven out of 140 detected metabolites, enriched in six metabolic pathways, were found significantly affected in E. coli metabolome. In summary, a targeted metabolomic platform is developed for high-throughput metabolite profiling and automated data processing, and the MRMAnalyzer program is a high efficient informatics tool for large scale targeted metabolomics.





Similar content being viewed by others
References
Bajad, S. U., Lu, W. Y., Kimball, E. H., Yuan, J., Peterson, C., & Rabinowitz, J. D. (2006). Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. Journal of Chromatography A, 1125(1), 76–88. doi:10.1016/j.chroma.2006.05.019.
Buescher, J. M., Moco, S., Sauer, U., & Zamboni, N. (2010). Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Analytical Chemistry, 82(11), 4403–4412. doi:10.1021/Ac100101d.
Chen, Q., Park, H. C., Goligorsky, M. S., Chander, P., Fischer, S. M., & Gross, S. S. (2012). Untargeted plasma metabolite profiling reveals the broad systemic consequences of xanthine oxidoreductase inactivation in mice. PLoS ONE, 7(6), e37149. doi:10.1371/journal.pone.0037149.
Clasquin, M. F., Melamud, E., & Rabinowitz, J. D. (2012). LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Current Protocols Bioinformatics, 14(Unit14), 11. doi:10.1002/0471250953.bi1411s37.
Dettmer, K., Aronov, P. A., & Hammock, B. D. (2007). Mass spectrometry-based metabolomics. Mass Spectrometry Reviews, 26(1), 51–78. doi:10.1002/Mas.20108.
Dudley, E., Yousef, M., Wang, Y., & Griffiths, W. J. (2010). Targeted metabolomics and mass spectrometry. Advances in Protein Chemistry and Structural Biology, 80, 45–83. doi:10.1016/S1876-1623(10)80002-1.
Fiehn, O. (2002). Metabolomics—The link between genotypes and phenotypes. Plant Molecular Biology, 48(1–2), 155–171. doi:10.1023/A:1013713905833.
Horai, H., Arita, M., Kanaya, S., Nihei, Y., Ikeda, T., Suwa, K., et al. (2010). MassBank: A public repository for sharing mass spectral data for life sciences. Journal of Mass Spectrometry, 45(7), 703–714. doi:10.1002/jms.1777.
Katajamaa, M., Miettinen, J., & Oresic, M. (2006). MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics, 22(5), 634–636. doi:10.1093/bioinformatics/btk039.
Kvitvang, H. F. N., Andreassen, T., Adam, T., Villas-Boas, S. G., & Bruheim, P. (2011). Highly sensitive GC/MS/MS method for quantitation of amino and nonamino organic acids. Analytical Chemistry, 83(7), 2705–2711. doi:10.1021/Ac103245b.
Lenz, E. M., & Wilson, I. D. (2007). Analytical strategies in metabonomics. Journal of Proteome Research, 6(2), 443–458. doi:10.1021/Pr0605217.
Locasale, J. W., Melman, T., Song, S., Yang, X., Swanson, K. D., Cantley, L. C., et al. (2012). Metabolomics of human cerebrospinal fluid identifies signatures of malignant glioma. Molecular & Cellular Proteomics, 11(6), M111 014688, doi:10.1074/mcp.M111.014688.
Lommen, A. (2009). MetAlign: Interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Analytical Chemistry, 81(8), 3079–3086. doi:10.1021/Ac900036d.
MacLean, B., Tomazela, D. M., Shulman, N., Chambers, M., Finney, G. L., Frewen, B., et al. (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics, 26(7), 966–968. doi:10.1093/bioinformatics/btq054.
Mazzarino, M., de la Torre, X., & Botre, F. (2008). A screening method for the simultaneous detection of glucocorticoids, diuretics, stimulants, anti-oestrogens, beta-adrenergic drugs and anabolic steroids in human urine by LC-ESI-MS/MS. Analytical and Bioanalytical Chemistry, 392(4), 681–698. doi:10.1007/s00216-008-2292-5.
Mezey, E., Dehejia, A., Harta, G., Papp, M. I., Polymeropoulos, M. H., & Brownstein, M. J. (1998). Alpha synuclein in neurodegenerative disorders: Murderer or accomplice? Nature Medicine, 4(7), 755–757. doi:10.1038/Nm0798-755.
Nicholson, J. K., & Lindon, J. C. (2008). Systems biology—Metabonomics. Nature, 455(7216), 1054–1056. doi:10.1038/4551054a.
Patti, G. J., Yanes, O., & Siuzdak, G. (2012). Metabolomics: The apogee of the omics trilogy. Nature Reviews Molecular Cell Biology, 13(4), 263–269. doi:10.1038/Nrm3314.
Pernet, C., Munoz, J., & Bessis, D. (2015). PENS (papular epidermal nevus with “skyline” basal cell layer). Annales de Dermatologie et de Venereologie, 142(1), 41–45. doi:10.1016/j.annder.2014.09.003.
Rabinowitz, J. D., & Silhavy, T. J. (2013). Systems biology: Metabolite turns master regulator. Nature, 500(7462), 283–284. doi:10.1038/nature12544.
Reiter, L., Rinner, O., Picotti, P., Huttenhain, R., Beck, M., Brusniak, M. Y., et al. (2011). mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nature Methods, 8(5), 430–435. doi:10.1038/nmeth.1584.
Smith, C. A., O’Maille, G., Want, E. J., Qin, C., Trauger, S. A., Brandon, T. R., et al. (2005). METLIN: a metabolite mass spectral database. Therapeutic Drug Monitoring, 27(6), 747–751.
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R., & Siuzdak, G. (2006). XCMS: Processing mass spectrometry data for metabolite profiling using Nonlinear peak alignment, matching, and identification. Analytical Chemistry, 78(3), 779–787. doi:10.1021/Ac051437y.
Tautenhahn, R., Bottcher, C., & Neumann, S. (2008). Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics,. doi:10.1186/1471-2105-9-504.
Tautenhahn, R., Cho, K., Uritboonthai, W., Zhu, Z. J., Patti, G. J., & Siuzdak, G. (2012). An accelerated workflow for untargeted metabolomics using the METLIN database. Nature Biotechnology, 30(9), 826–828. doi:10.1038/Nbt.2348.
Tautenhahn, R., Patti, G. J., Kalisiak, E., Miyamoto, T., Schmidt, M., Lo, F. Y., et al. (2011). metaXCMS: second-order analysis of untargeted metabolomics data. Analytical Chemistry, 83(3), 696–700. doi:10.1021/ac102980g.
Tsugawa, H., Arita, M., Kanazawa, M., Ogiwara, A., Bamba, T., & Fukusaki, E. (2013). MRMPROBS: A data assessment and metabolite identification tool for large-scale multiple reaction monitoring based widely targeted metabolomics. Analytical Chemistry, 85(10), 5191–5199. doi:10.1021/Ac400515s.
Tsugawa, H., Kanazawa, M., Ogiwara, A., & Arita, M. (2014). MRMPROBS suite for metabolomics using large-scale MRM assays. Bioinformatics, 30(16), 2379–2380. doi:10.1093/bioinformatics/btu203.
Wei, R., Li, G. D., & Seymour, A. B. (2010). High-throughput and multiplexed LC/MS/MRM method for targeted metabolomics. Analytical Chemistry, 82(13), 5527–5533. doi:10.1021/Ac100331b.
Wong, J. W., Abuhusain, H. J., McDonald, K. L., & Don, A. S. (2012). MMSAT: Automated quantification of metabolites in selected reaction monitoring experiments. Analytical Chemistry, 84(1), 470–474. doi:10.1021/ac2026578.
Xia, J. G., Psychogios, N., Young, N., & Wishart, D. S. (2009). MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Research, 37, W652–W660. doi:10.1093/Nar/Gkp356.
Xia, J. G., & Wishart, D. S. (2011). Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nature Protocols, 6(6), 743–760. doi:10.1038/nprot.2011.319.
Yuan, M., Breitkopf, S. B., Yang, X. M., & Asara, J. M. (2012). A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nature Protocols, 7(5), 872–881. doi:10.1038/nprot.2012.024.
Zhu, Z. J., Schultz, A. W., Wang, J. H., Johnson, C. H., Yannone, S. M., Patti, G. J., et al. (2013). Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nature Protocols, 8(3), 451–460. doi:10.1038/nprot.2013.004.
Acknowledgments
We thank the financial support provided by the startup funding from Interdisciplinary Research Center on Biology and Chemistry (IRCBC), Chinese Academy of Sciences (CAS). Z.-J. Z. is also supported by Thousand Youth Talents Program (The Recruitment Program of Global Youth Experts from Chinese government). The project is also financially supported by Agilent Technologies Thought Leader Award.
Compliance with ethical requirements
All institutional and national guidelines for the care and use of biological samples were followed. The data acquired were in accordance with appropriate ethical requirements.
Conflict of interest
There are no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Yuping Cai and Kai Weng have contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cai, Y., Weng, K., Guo, Y. et al. An integrated targeted metabolomic platform for high-throughput metabolite profiling and automated data processing. Metabolomics 11, 1575–1586 (2015). https://doi.org/10.1007/s11306-015-0809-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-015-0809-4