Abstract
Microsatellite markers are still the marker of choice for many research questions in the field of forest genetics. However, the number of available markers is often low for species that have not been studied intensively like the tree of heaven (Ailanthus altissima). During the last decade, next-generation sequencing (NGS) has offered advanced techniques for efficiently identifying microsatellite markers and accurately genotyping samples. Here, we identify new microsatellite markers for the tree of heaven by applying an NGS-based method using the Illumina MiSeq platform. NGS technology was proved to be an effective method for fast and cost-efficient identification of microsatellite markers by implementing a genotyping-by-sequencing approach based on Illumina amplicon sequencing (SSR-GBS). We screened three populations from Eastern Austria for genetic variation at 19 newly identified microsatellite loci. We tested two different genotyping approaches: (1) considering only allele lengths (forming a so-called “allele length dataset”), (2) taking also single nucleotide polymorphisms (SNPs) within the amplified fragments into account (forming a so-called “SNP dataset”). The results revealed higher values for all genetic diversity parameters, as well as a better resolution of genetic assignment, when the latter approach was followed. Thus, by taking advantage of sequence information which is provided by SSR-GBS, one may achieve considerable gains in performance using the same marker set. The developed markers provide a cost-efficient tool for genotyping populations of tree of heaven and the approach presented here promises to be of high value for medium throughput genotyping applications in non-model forest tree species. We will use this method to widen the perspectives for further population genetic investigations of the tree of heaven.
Similar content being viewed by others
Introduction
Microsatellites, consisting of short tandem repetitive DNA motifs that range from one to six nucleotides in length (also known as simple-sequence repeats (SSR) or short tandem repeats (STR)), have been one of the most widely used molecular markers (Guichoux et al. 2011; de Barba et al. 2017). They have played an important role in identifying population genetic structure of numerous species and have been applied in parentage analysis, conservation ecology, phylogenetic analysis and population genetic studies (Aldrich et al. 1998; Khasa et al. 2003; Addisalem et al. 2015; Zhao et al. 2015). In comparison to other markers, such as single nucleotide polymorphisms (SNPs), the major advantages of microsatellites are the higher statistical power per locus, higher allelic richness and higher mutation rate (Farrell et al. 2016).
Before the emergence of next-generation sequencing (NGS) technologies, the development of microsatellite markers was mostly based on microsatellite enrichment. This method involves several steps like genomic DNA digestion with restriction enzymes, hybridization of oligonucleotides, and cloning (Karagyozov et al. 1993; Gardner et al. 1999; Dallas et al. 2005). From the current point of view, such procedures are time-consuming, laborious, and expensive, particularly for species being examined for the first time (Honig et al. 2017). Identifying SSRs has become cheaper, faster, and easier to implement with the emergence of NGS technologies. Briefly, these methods consist in (i) identification of repetitive motifs (microsatellites) using genome-wide sequence data produced using a NGS-method, and (ii) designing primers for amplifying the targeted loci. Nowadays, there are several examples demonstrating how these technologies, mostly Illumina, can efficiently identify microsatellite markers (Cai et al. 2013; Castoe et al. 2012; Farrell et al. 2016; Qin et al. 2017).
Use of NGS has also brought about improvements in SSR-scoring. Traditionally, SSRs have been genotyped by assessing fragment length variation through capillary electrophoresis, as a proxy for the number of repetition units (Guichoux et al. 2011). However, sequence length variation alone might miss some useful information. Furthermore, it is prone to scoring errors caused by PCR amplification artifacts and suffers from imprecise sizing, length homoplasy and limited ability to multiplex multiple loci per sample (Darby et al. 2016). These limitations can be overcome by applying NGS techniques and genotyping microsatellites using allele calling based on sequence information (henceforth called SSR-genotyping-by-sequencing; SSR-GBS). With this approach, it is possible to include SNP information, resulting in a higher number of alleles and, thus, increase the potential statistic power of the SSR marker (de Barba et al. 2017; Tibihika et al., Genetic structure of the anthropogenically threatened East African Oreochromis niloticus; Linnaeus 1758: Towards a conservation pipeline based on SSR-genotyping-by-sequencing (GBS), submitted). At the same time, size homoplasy is prevented, which is due to alleles identical by length, but not by sequence (Selkoe and Toonen 2006). Other advantages of SSR-GBS are a higher automation, replicability and a lower cost of the genotyping process (de Barba et al. 2017; Vartia et al. 2016; Zhan et al. 2017). Automation and replicability are achieved by using available bioinformatics tools and simple scripts, and by preventing the artifacts associated with fragment analysis. Such improvements allow for the standardization of SSR genotyping (Farrell et al. 2016; de Barba et al. 2017), something that was impossible with the traditional electropherogram sequence-length procedures.
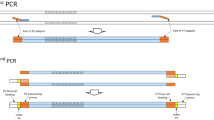
Furthermore, SSR-GBS combined with Illumina amplicon sequencing allows for an increased efficiency which can be achieved by sequencing multiplexed PCR products (i) from different individuals and (ii) from a much higher number of loci, compared to the multiplexing possibilities based on fragment length scoring (Guichoux et al. 2011). Multiplex sequencing can be implemented by tagging the target DNA fragments from different samples with different DNA barcodes (Elshire et al. 2011). In Illumina sequencing platform, this is mostly done through the incorporation of indexes which are added during preparation of the sequencing libraries via PCR reactions or ligation of adaptors (Shokralla et al. 2015; Meimberg et al. 2016; de Barba et al. 2017).
In the present study, we use an SSR-GBS approach using an Illumina-MiSeq platform to develop and analyze microsatellite loci for the tree of heaven (Ailanthus altissima (Mill.) Swingle), a tree species with a very limited number of molecular markers developed thus far. First, we identify new SSR loci for this species by using NGS (Illumina MiSeq platform) and design primers for amplifying the respective loci. Second, we test amplification and variability of these loci in population samples by co-amplifying multiple microsatellites in multiplex PCR reactions, adding individual tags and pooling all PCR products simultaneously in a single flow-cell lane of the Illumina sequencing platform. Third, we compare the gain in information by accessing amplicon sequence information rather than only length information, i.e., SNPs and insertions-deletions.
Materials and methods
Study populations and DNA isolation
We used one sample collected from a tree of heaven in the BOKU University premises in Vienna for microsatellite discovery. 20 ng of genomic DNA (gDNA) were isolated from leaf tissue using magnetic bead DNA isolation technology implemented in the Plant Kit (Magneamedics) following the manufacturer’s recommendations. The concentration of the isolated DNA was evaluated on a 1.8% agarose gel stained with HDGreen®Plus Safe DNA Dye (INTAS); fragments were visualized in ultraviolet light. The isolated gDNA was submitted for whole genome low coverage sequencing to an Illumina MiSeq sequencing run (Illumina) in the Ludwig-MaximilianUniversity of Munich, Germany.
Based on previous research (Pötzelsberger and Hasenauer 2015) and information provided by local forest managers, three study stands with tree of heaven were selected for genetic analysis (Fig. 1). All three study stands represent forest sites invaded by the tree of heaven about three (plots Brunnkirchen and Donnerskirchen; Fig. 1) to six (plot Matzen; Fig. 1) decades ago. No plantations took place. In all cases, old individuals of heaven tree are present in nearby villages and these might have served as seed sources for the colonization of the study plots (personal communication with local forest managers).
Collection of plant tissue for genetic analysis was conducted according to the following specific sampling design: a reference grid of 20 m × 20 m was established within each forest stand and plant tissue (cambium or buds) from 30 trees with a diameter at breast height (dbh) ≥ 7 cm was collected (90 trees in total). After sampling, plant leaf tissues were put into a plastic bag with silica gel and stored at room temperature for drying. DNA was extracted using the DNEasy Plant Minikit (Qiagen, Hilden, Germany).
Microsatellite discovery and primer development
After receiving the sequence reads from the whole genome sequencing carried out with the sample from the BOKU university premises, we applied trimming, quality filtering, read merging, and identified microsatellite motifs. In particular, low-quality read regions and adapters were trimmed and filtered, using the Cutadapt (Martin 2011) program. Read 1 and Read 2 were merged using the program PEAR (Zhang et al. 2013). We used the program SSR_pipeline (Miller et al. 2013) to select reads containing microsatellite motifs that passed the following criteria: 40 bp flanking on both sides of the motif; at least six repetitions for tetra- and pentanucleotide motifs, and at least 10 and 8 repetitions for di- and trinucleotides motifs, respectively. Primer design was carried out using the Primer 3 Plus program (Untergasser et al. 2007) implemented as a plugin in the Geneious v. 8.1.8 program (Kearse et al. 2012) with the following requirements: optimal melting temperature of 52–59 °C, a GC content ranging from 20 to 80%, an optimal oligo length between 18 and 21 base pairs, and the amplification product size from 350 to 450 base pairs (in order to be suitable for sequencing based on the Illumina MiSeq).
PCR amplification
Marker performance validation
For marker validation, PCR reactions were carried out using the samples from the three forest stands. Each reaction had a total volume of 10 μl containing 5 μl QIAGEN Multiplex PCR Kit (QIAGEN), 4 μl primer solution (each 1 μM forward and reverse), 0.6 μl H2O, and 0.4 μl original genomic DNA used for primer design. The PCR was performed using the following steps: initial denaturation at 95 °C for 15 min followed by 35 cycles of denaturation at 95 °C for 30 s, primer annealing at 55 °C for 1 min, and extension at 72 °C for 1 min followed by a final extension of 10 min.
The designed primer pairs were pooled into three primer mixes with diluted concentration of 3 μM for each forward and reverse primer. All three primer mixes were tested for multiplex reactions using original genomic DNA which we used for primer design.
Multiplex and index PCRs
DNA libraries were prepared in two PCR steps. In a first PCR, specific primers, extended with parts of the Illumina P5 (TCTTTCCCTACACGACGCTCTTCCGATCT,) and P7 (CTGGAGTTCAGACGTGTGCTCTTCCGATCT) adapters were used for amplification of the respective marker. In the second PCR, the Illumina adapters were added including the index information which allows sample identification. This strategy resulted in combinations of two 8-bp-long DNA sequences (P5 AATGATACGGCGACCACCGAGATCTACAC[Index]ACACTCTTTCCCTACACGACG; P7: CAAGCAGAAGACGGCATACGAGAT[Index]GTGACTGGAGTTCAGACGTGT). PCR multiplex reactions were employed in a total volume of 10 μl contained with 5 μl of the QIAGEN Multiplex PCR Kit (QIAGEN), 0.5 μl primer mix (1 μM), 3.5 μl H2O, and 1 μl of undiluted DNA. The following protocol was applied: initial denaturation at 95 °C for 15 min followed by 35 cycles of 30 s of 95 °C, 55 °C for 1 min, and extension at 72 °C for 1 min followed by 10 min. PCR products from the same sample but different primer mixes were pooled together and purified (description below).
The second PCR reaction incorporated the index information and Illumina flow-cell binding sites. Reaction was carried out in a 10 μl total volume consisting of 5 μl QIAGEN Multiplex PCR Kit (QIAGEN), 2 μl P5 and P7 primers (1 μM) and 1 μl of the elution buffer from the clean-up reaction. PCR reactions included following steps: initial denaturation of 95 °C for 8 min, followed by 10 cycles of 95 °C for 30 s, 58 °C for 1 min 72 °C for 1 min followed by 5 min. PCRs were performed using a BIO-RAD T100tm Thermal Cycler. Quality of all amplified PCR products were visualized on 2% agarose gel stained with HDGreen®Plus Safe DNA Dye (INTAS) and fragments visualized in ultraviolet light.
PCR purification
After each PCR step, the resulting products were cleaned to remove possible primer dimers and unused primers. Before clean-up, we pooled the products from the three PCR multiplex reactions. We took 1 μl from the primer mixes M1 and M3 and 2 μl from the M3 (Table 1). The latter primer mix resulted in a lower amount of PCR product than the others. It was for this reason that we added the double amount (2 µl) from this mix (M3) to keep an equal representation of the three mixes in the pooled solution. Pooling was done with the purpose of saving time and costs. From each pooled PCR product, 4 μl were mixed with 2.8 μl magnetic bead solution (Steinbrenner) and incubated for 5 min. PCR products now bound to the magnetic beads were transferred for the next steps using a Magnetic Bead Extraction Replicator VP 407-AM-N inverse magnet using manufacturer specifications (V&P Scientific, INC., San Diego, CA 92121, USA). Washing was done twice by dipping the beads into 200 μl of 80% ethanol for 45 s. For the next step they were dried at room temperature for 5 min on the magnet holder. In a last step, PCR products were eluted with the 17 μl elution buffer (65 °C).
After generating libraries, 1.5 μl of all indexed and purified PCR products was pipetted together into a new 1.5-ml Eppendorf tube and sent for an Illumina MiSeq run at the Genomics Service Unit from the Ludwig-Maximilian University, Munich, Germany. We used 6% of the MiSeq run expecting to recover 837,900 reads and a coverage per marker per sample of 266x.
Sequence data analysis
Raw data processing after Illumina MiSeq run
Both sequence processing and allele calling (next paragraph) were done using several bash, python and R scripts that are listed in Tibihika et al., Genetic structure of the anthropogenically threatened East African Oreochromis niloticus; Linnaeus 1758: Towards a conservation pipeline based on SSR-genotyping-by-sequencing (GBS), submitted) and are available in github (https://github.com/mcurto/SSR-GBS-pipeline). Sequences were filtered for quality control and merged using the program PEAR (Zhang et al. 2013) using the default parameters (Martin 2011). Only those merged reads were retained for further analysis, which contained primer sequences on both sides (starting with forward and ending with reverse). Moreover, a python script and a primer file split up reads from different primers in separate files.
Genotyping
Genotyping was performed in two steps. First, alleles were defined according to sequence length. Read lengths per sample and marker were obtained by using an awk script, which was then used to create read length statistics as input for a custom R script. The following criteria were used for distinguishing between homozygous, heterozygous, and ambiguous genotypes: homozygote genotypes contained only one dominant read length comprising a minimum user defined alpha value (here: 62% of all reads), whereas heterozygous genotypes possessed two most frequent read lengths, which together contained more than 62% of all reads (Tibihika et al., Genetic structure of the anthropogenically threatened East African Oreochromis niloticus; Linnaeus 1758: Towards a conservation pipeline based on SSR-genotyping-by-sequencing (GBS), submitted; Winter 2018). Additional criteria were set to distinguish alleles from stutter bands or to account for the differential amplification of alleles: e.g., if the second largest allele was one repeat motif length smaller than the most abundant one, its relative abundance needed to exceed 75% of the most abundant allele. In case these criteria were not met, the sample was marked for manual control. The R script produced one co-dominant csv matrix file and one pdf containing all length histograms per marker and sample to facilitate visualization for manual control. Alleles were assigned a number corresponding to the sequence length.
In a second step, we considered SNP variation and combined it with length information for the final allele call. In this way, consensus sequences with the same length but differences at SNP-loci were treated as different alleles. For this purpose, the reads corresponding to the alleles called in the first step were extracted and a 70% consensus sequence per allele (according to its length inferred previously) and sample was created. This means that, for each position, the respective base was retained as consensus if its frequency was above 70%. If none of the four nucleotides met this criterion, we considered this position as a potential heterozygote SNP. The variants from each potential SNP were obtained by screening the reads used to make the consensus sequence and recovering the nucleotide frequencies for each nucleotide in the SNP position. The two most frequent nucleotides were recovered and used to separate the consensus sequence into two. In case more than one potential SNP was found, we looked for the two most frequent linked variants among several positions. For example, in case of the potential SNPs A/T at position 55 and A/C at position 200, we would look for the frequency of A(55)A(200), A(55)C(200), T(55)A(200), and T(55)C(200), and used the two most frequent combinations to separate the consensus sequence. If ambiguous bases were found in a heterozygote length genotype, they were corrected using only the most frequent nucleotide variant. Finally, each unique sequence per locus was assigned a unique numeric allele name. These allele names were used to call genotypes, which were saved in a co-dominant matrix.
Monomorphic loci and loci with more than 50% missing genotypes (presumably due to null alleles caused by mutations at the primer regions which hinder primer hybridization; Pemberton et al. 1995) were not included in the population genetic analysis. A further criterion to exclude markers from the analysis was the presence of too many potential alleles (i.e., many different sequences with similar coverage), which did not allow scoring of one or two alleles per individual. These were also discarded because they probably stem from unspecific products or duplicated loci.
Population genetic analysis
After narrowing down our marker selection according to the aforementioned criteria, we performed population genetic analyses using genotypic data from the three forest stands mentioned above. First, we used the GenAlEx software (Peakall and Smouse 2006, 2012) to compute the number of alleles, as well as the observed and expected heterozygosity per locus and over all loci. Second, we calculated inbreeding coefficients (FIS according to Weir and Cockerham 1984) and pairwise fixation indices (FST values; Weir and Cockerham 1984) using the FSTAT-software (Goudet 1995). We tested significance of these coefficients by performing the maximum number of possible randomizations of alleles among individuals within samples for FIS and of genotypes among populations for FST. No Hardy-Weinberg Equilibrium was assumed. Third, we performed a Bayesian cluster analysis using the STRUCTURE software (Pritchard et al. 2000; Falush et al. 2003) in order to investigate genetic structure across the sampled populations. For the STRUCTURE analysis, we chose the admixture model and correlated allele frequencies. We set the number of assumed subpopulations/clusters (K) between 1 and 10. For each K, we performed 20 independent runs applying 50,000 burn-in replications followed by 100,000 MCMC iterations. STRUCTURE analysis was carried out with the program STRAUTO v1.0 which allows automation and parallelization of multiple STRUCTURE runs (Chhatre and Emerson 2017). We used the web server CLUMPAK (Kopelman et al. 2015) to align the inferred clusters and average membership proportions across runs within each K-value. In order to detect the uppermost hierarchical level of structure, we assessed the parameter ΔK after Evanno et al. (2005) using the software Structure-Harvester (Earl 2012). Fourth, we calculated inter-individual genetic distances according to Huff et al. (1993) in GenAlEx and performed a principal coordinate analysis (PCoA; Orlóci 1978) based on these distances.
All population genetic analyses were carried out (i) using the dataset with allele lengths, henceforth called “allele length dataset,” and (ii) using the dataset considering both SNP and allele length variation, henceforth called “SNP dataset.” By repeating the analyses within each one of the datasets, we aimed to compare (i) the discrepancy of the genetic diversity estimates and (ii) a possible difference in the resolution of genetic structures.
In addition, all multilocus analyses were carried out using a reduced marker set which included only the markers which did not present any unspecific bands at all, as mentioned above.
Results
Screening for microsatellites, primer development, and marker validation
The NGS run of the sample used for microsatellite discovery resulted in 1,174,986 reads. After filtering and merging, 985,001 reads were retained. The SSR pipeline detected 2084 sequences with microsatellite motifs (820 di-, 1005 tri-, 218 tetra- and 41 penta-nucleotides). From these sequences, primer pairs for the amplification of 13 di-, 21 tri-, and two tetra-nucleotide microsatellites were designed (Table 1). All 36 primer pairs successfully amplified from the single original Ailanthus altissima sample that we used for the primer design. However, when multiplex PCR was applied using individuals from forest populations, four loci could not be amplified (HT20, HT25, HT33, and HT34). For genotyping, we obtained a total of 960,701 paired reads which were reduced to 875,836 after quality filtering and demultiplexing. The number of reads per marker and sample varied between 0 and 2062 with an average coverage of 270×. The overall amount of missing data in the matrix was 31.9%, which was reduced to 5.1% after dicarding a total of eight lower quality markers due to missing data (Table 1). Nineteen of the remaining 32 loci fulfilled our criteria regarding polymorphism and success of amplification, as shown by the genotyping of forest stands (Table 1). These were used for population genetic analyses. The primers for these loci are shown in Table 1. Six among these 19 loci (HT12, HT13, HT15, HT17, HT21, and HT32) presented non-specific products or stutter bands, which hindered automatic scoring. For this reason, the scoring process was done manually for these six markers. Because stutter bands may have introduced some errors is the scoring, we repeated all multilocus population genetic analyses by excluding these six loci, i.e., by including 13 loci that were scored completely unambiguously. Finally, during genotyping of locus HT35, we ignored one additional PCR-product (length “360”), which occurred in all samples. This locus was retained for all steps of population genetic analysis.
Genetic diversity over loci and populations
Mean number of alleles (Na), observed heterozygosity (Ho), expected heterozygosity (He), and mean inbreeding coefficient per locus and standard error (FIS) differed between the “allele length dataset” and the “SNP dataset”. Higher values for the diversity parameters (Na, Ho, He) were found using the “SNP dataset” (Table 2). Using the “allele length dataset,” number of alleles (Na) varied between 2 (loci HT15 and HT30) and 8 (locus HT17 and HT32) with a mean of 4.58 and a standard error of 0.41 across loci and populations (Table 2). For the “SNP dataset,” the number of alleles per marker varied between 3 (locus HT19) and 17 (locus HT29), displaying a mean and standard error of 6.58 and 0.74, respectively (Table 2). For eight loci, there was not a significant difference between observed and expected heterozygosity (i.e., significant FIS-values), either when values were calculated based on the “allele length dataset” or the “SNP dataset” (Table 2). With the exception of locus HT13 and HT32, all loci displaying non-specific PCR products or stutter bands were characterized by a significant heterozygote deficit (Table 2). Some of these loci (e.g., HT15 and HT17) also exhibited a high number of alleles (Na). Thus, removal of those loci resulted in a lower mean number of alleles per locus, whereas other values of genetic differentiation remained unchanged (Table 2).
Genetic differentiation and population genetic structure
Pairwise genetic differentiation in terms of FST was high and significant between almost all population pairs. In particular, a non-significant FST = 0.07 was found between Matzen and Donnerskirchen when the “SNP dataset” with all 19 markers was used. All other pairwise comparisons were significant. Pairwise FST values varied between 0.044 and 0.096 and all of them were somewhat higher when computed with the “SNP dataset” (Table 3a, b).
Bayesian cluster analysis with STRUCTURE displayed some differences depending on the used dataset. For three datasets (“allele length” and “SNP dataset” with 19 loci and “allele length dataset” with 13 loci), the ΔΚ-statistic was maximized for K = 2, suggesting two subpopulations as the uppermost level of population clustering. For the ‘SNP dataset’ with 13 loci, ΔK was maximized at K = 3 (Online Resource 1). Similar patterns were revealed for K = 2 in all four cases when inspecting the bar plots with the individual membership proportions to the modeled clusters: populations Donnerskirchen and Matzen formed a common cluster, while most individuals from Brunnkirchen were assigned to the other cluster. At K = 3, three clusters were resolved corresponding to the three populations (Fig. 2). However, the “SNP dataset” resulted in a higher membership proportion of individuals to their own population. Analysis using the “allele length dataset” and only 13 loci did not successfully assign Donnerskirchen and Matzen populations to different clusters (Online Resource 2). No further meaningful structure was revealed at K = 4. In general, membership proportions of individuals to their own population cluster were higher when the “SNP dataset” was used.
The ordination of inter-individual genetic distances by means of a PCoA resulted in different groups for each of the study populations with varying degree of overlapping, depending on the dataset used (Fig. 3). In general, populations could be distinguished better when the “SNP dataset” was used. The number of loci did not have any obvious influence on the degree of overlapping between populations (Fig. 3, Online Resource 2).
Ordination along the first and second principal coordinates derived by a principal coordinate analysis (PCoA) based on interindividual distances using the “allele length dataset” (above) and the “SNP dataset” (below) based on genotypic data from 19 markers. Each point represents an individual. Populations are depicted with different shapes/colors. The percentage of explained variation by the first two coordinates was 19.0 and 18.2% respectively
Discussion
With our study, we provide a new set of molecular markers for population genetic analyses of tree of heaven and present an efficient approach for describing patterns of genetic differentiation. By integrating Illumina amplicon sequencing with microsatellite genotyping, researchers can achieve a higher throughput and reproducibility, and increase the statistical power (Vartia et al. 2016; de Barba et al. 2017; Tibihika et al., Genetic structure of the anthropogenically threatened East African Oreochromis niloticus; Linnaeus 1758: Towards a conservation pipeline based on SSR-genotyping-by-sequencing (GBS), submitted). This last effect results in a higher number of alleles, which are obtained by the ability of recovering sequence information combined with the microsatellite length. The additional sequence information can also help to resolve length homoplasy in microsatellites, as we demonstrate here. For most markers, we found a higher number of alleles when we took SNPs into account (see Table 2). The high mutation rate that results in this high number of alleles also results in alleles of different origin (i.e., not identical by descent) but with the same length (Estoup et al. 2002). By including sequence information (use of “SNP dataset”), some of these cases can be differentiated reducing this effect and increasing marker diagnostic power.
Indeed, by accounting for SNP information, we achieved a higher resolution for describing genetic differentiation. For instance, using the “SNP dataset” and assuming three subpopulations (K = 3), we could differentiate well among the three study populations and most individuals showed a high membership proportion to their own population cluster, even when using a reduced number of 13 loci. This was not the case for the “allele length dataset” and 13 markers, which performed worse for K = 3. In the latter case, populations Donnerskirchen and Matzen were shown as admixed, with more or less equal membership proportions (ca. 0.4–0.5) to each one of two clusters. In addition, when accounting for sequence information, we found higher values of pairwise FST and we could better differentiate between the three populations based on individual genotypes . In general, consideration of SNP information resulted in a moderate increase of allele number, which is due to the detection of alleles being identical by fragment length, but not by sequence information (e.g., loci HT2, HT7, HT9 in Table 2). In our case, adding SNP information has a higher impact than increasing the number of loci. We expect that saturation in amount of information obtained is achieved with an increasing number of markers to the point where increasing the number of alleles per marker should not have an effect.
There were two cases (loci HT15 and HT17) with a strong increase of allele number when SNPs were taken into account (Table 2). Here, the differences could also be a consequence of a faulty SNP call due to stutter band artifacts: in case of a heterozygote length-based genotype with a SNP between both alleles, stutter bands of the larger allele would insert SNP variation present in the larger allele into the smaller allele. This could create erroneous genotypes, especially if the markers were sequenced with low coverage or if the primers are more efficiently amplifying one allele over the other. The datasets consisting of 13 markers were free of such ambiguities. Reducing the number of loci from 19 to 13 only had a minimal effect on the results on diversity and differentiation among the three study populations.
Due to missing data, we ended up excluding eight markers out of the initial marker set of markers tested (see Table 1). The main source of amplification failure was likely to be related with the plexity level of the PCR reactions and not due to the SSR-GBS approach. Evidence of this is the fact that all markers amplified when a single marker PCR approach was employed. In our approach, we choose the strategy of developing more markers than necessary and exclude the ones that could not be amplified in multiplex or presented scoring problems. However, by reducing the number of markers per multiplex or optimizing the PCR reactions, more markers are likely to be recovered. PCR reactions with similar plexity are commonly successfully applied for forensics purposes (Heyen et al. 1997; Luikart et al. 1999).
Our SSR-GBS approach also provides a cheaper alternative compared to traditional length analyses methods. For example, considering 32 markers, a PCR multiplex of four markers per reaction, and a sequencing coverage of 600, we estimate that SSR-GBS would require around 6 € per sample (Online Resource 3). To recover the same data, traditional SSR genotyping would cost around 26 € per sample (Online Resource 3). The main reason for this cost discrepancy is the ability of multiplexing all markers after the first PCR reaction given the fact that the obtained sequencing data can be later demultiplexed independently of the number of markers used. This does not apply for fragment length analysis where the number of markers per capillary run are restricted by the number of florescent dyes used. Another contributor is the relatively low sequence cost of the Illumina platforms. One Illumina MiSeq run is estimated to produce 15 million paired reads with a price rounding 1600 euros. Considering a coverage of 600× this would result on a sequencing cost of 0.064 € per marker per sample.
In previous studies, other authors focused on further advantages of using high-throughput sequencing technologies for microsatellite genotyping. For example, de Barba et al. (2017) highlighted the reproducibility of this method (especially by avoiding errors due to subjective binning, which was shown to contribute to 83% of all discrepancies between laboratories; Weeks et al. 2002), and other studies such as Vartia et al. (2016) and Farrell et al. (2016) demonstrated the high throughput of the method. Among these characteristics, the high reproducibility will contribute to the application of microsatellites in large long-term projects, and combination of multiple datasets produced by independent researchers. Given these characteristics together with the higher statistical power, we expect that this method will replace the capillary electrophoresis approach and prolong the relevance of microsatellites in the genomic era. Many studies have limited budgets or comprise questions that do not require whole genome data but rather the genotyping of many individuals. For these reasons, microsatellites are still the markers of choice.
So far, in the case of Ailanthus altissima, available markers from its nuclear genome include nine microsatellites developed by Dallas et al. (2005). These loci or a subset of them have been used for analyses of population genetic structure (Aldrich et al. 2010; Kurokochi et al. 2014) or clonality (Chuman et al. 2015). However, depending on the scientific question, a population genetic study may require a higher number of markers. For instance, an increased number of loci results in a higher accuracy for identifying ramets of the same clone, while presence of half- or full-sibs increases the need of a larger marker set required for this purpose (Wang 2016). Given that the tree of heaven has successfully colonized large areas outside its native range (Kowarik and Säumel 2007), it will be of particular interest to perform population genetic studies in this species in order to understand the genetics of biological invasions. We expect that the new set of highly variable markers presented here, as well as the increased resolution of our genotyping approach, will widen the possibilities of studying population genetics of the tree of heaven.
References
Addisalem AB, Esselink GD, Bongers F, Smulders MJ (2015) Genomic sequencing and microsatellite marker development for Boswellia papyrifera, an economically important but threatened tree native to dry tropical forests. AoB Plants 7:plu086
Aldrich PR, Hamrick JL, Chavarriaga P, Kochert G (1998) Microsatellite analysis of demographic genetic structure in fragmented populations of the tropical tree Symphonia globulifera. Mol Ecol 7(8):933–944
Aldrich PR, Briguglio JS, Kapadia SN, Morker MU, Rawal A, Kalra P, Huebner CD, Greer GK (2010) Genetic structure of the invasive tree Ailanthus altissima in eastern United States cities. J Bot 2010:795735
Cai G, Leadbetter CW, Muehlbauer MF, Molnar TJ, Hillman BI (2013) Genome-wide microsatellite identification in the fungus Anisogramma anomala using Illumina sequencing and genome assembly. PLoS One 8(11):e82408
Castoe TA, Poole AW, de Koning AJ, Jones KL, Tomback DF, Oyler-McCance SJ, Fike JA, Lance SL, Streicher JW, Smith EN, Pollock DD (2012) Rapid microsatellite identification from Illumina paired-end genomic sequencing in two birds and a snake. PLoS One 7(2):e30953
Chhatre VE, Emerson KJ (2017) StrAuto: automation and parallelization of STRUCTURE analysis. BMC Bioinf 18(1):192
Chuman M, Kurokochi H, Saito Y, Ide Y (2015) Expansion of an invasive species, Ailanthus altissima, at a regional scale in Japan. J Ecol Environ 38:47–56
Dallas JF, Leitch MJ, Hulme PE (2005) Microsatellites for tree of heaven (Ailanthus altissima). Mol Ecol Resour 5(2):340–342
Darby BJ, Erickson SF, Hervey SD, Ellis-Felege SN (2016) Digital fragment analysis of short tandem repeats by high-throughput amplicon sequencing. Ecol Evol 6(13):4502–4512
de Barba M, Miquel C, Lobréaux S, Quenette PY, Swenson JE, Taberlet P (2017) High-throughput microsatellite genotyping in ecology: improved accuracy, efficiency, standardization and success with low-quantity and degraded DNA. Mol Ecol Resour 17(3):492–507
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6(5):e19379
Estoup A, Jarne P, Cornuet JM (2002) Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol Ecol 11(9):1591–1604
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164(4):1567–1587
Farrell ED, Carlsson JE, Carlsson J (2016) Next Gen Pop Gen: implementing a high-throughput approach to population genetics in boarfish (Capros aper). R Soc Open Sci 3(12):160651
Gardner MG, Cooper SJB, Bull CM, Grant WN (1999) Isolation of microsatellite loci from a social lizard, Egernia stokesii, using a modified enrichment procedure. J Hered 90(2):301–304
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86(6):485–486
Guichoux E, Lagache L, Wagner S, Chaumeil P, Léger P, Lepais O, Lepoittevin C, Malausa T, Revardel E, Salin F, Petit RJ (2011) Current trends in microsatellite genotyping. Mol Ecol Resour 11(4):591–611
Heyen DW, Beever JE, Da Y, Everts RE, Green C, Lewin HA, Bates SRE, Ziegle JS (1997) Exclusion probabilities of 22 bovine microsatellite markers in fluorescent multiplexes for semiautomated parentage testing. Anim Genet 28(1):21–27
Honig JA, Zelzion E, Wagner NE, Kubik C, Averello V, Vaiciunas J, Bhattacharya D, Bonos SA, Meyer WA (2017) Microsatellite identification in perennial ryegrass using next-generation sequencing. Crop Sci 57(Supplement 1):S331–S340
Huff DR, Peakall R, Smouse PE (1993) RAPD variation within and among natural populations of outcrossing buffalograss [Buchloe dactyloides (Nutt.) Engelm.]. Theor Appl Genet 86(8):927–934
Karagyozov L, Kalcheva ID, Chapman VM (1993) Construction of random small-insert genomic libraries highly enriched for simple sequence repeats. Nucleic Acids Res 21(16):3911–3912
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649
Khasa DP, Nadeem S, Thomas B, Robertson A, Bousquet J (2003) Application of SSR markers for parentage analysis of Populus clones. For Genet 10(4):273–281
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15(5):1179–1191
Kowarik I, Säumel I (2007) Biological flora of central Europe: Ailanthus altissima (Mill.) swingle. Perspect Plant Ecol Evol Syst 8(4):207–237
Kurokochi H, Saito Y, Ide Y (2014) Genetic structure of the introduced heaven tree (Ailanthus altissima) in Japan: evidence for two distinct origins with limited admixture. Botany 93(3):133–139
Luikart G, Biju-Duval M-P, Ertugrul O, Zagdsuren Y, Maudet C, Taberlet P (1999) Power of 22 microsatellite markers in fluorescent multiplexes for parentage testing in goats (Capra hircus). Anim Genet 30(6):431–438
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17(1):10–12
Meimberg H, Schachtler C, Curto M, Husemann M, Habel JC (2016) A new amplicon based approach of whole mitogenome sequencing for phylogenetic and phylogeographic analysis: an example of East African white-eyes (Aves, Zosteropidae). Mol Phylogenet Evol 102:74–85
Miller MP, Knaus BJ, Mullins TD, Haig SM (2013) SSR_pipeline: a bioinformatic infrastructure for identifying microsatellites from paired-end Illumina high-throughput DNA sequencing data. J Hered 104(6):881–885
Orlóci L (1978) Multivariate analysis in vegetation research. Springer Netherlands, The Hague
Peakall RO, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Resour 6(1):288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research - an update. Bioinformatics 28:2537–2539
Pemberton JM, Slate J, Bancroft DR, Barrett JA (1995) Nonamplifying alleles at microsatellite loci: a caution for parentage and population studies. Mol Ecol 4(2):249–252
Pötzelsberger E, Hasenauer H (2015) High mortality in tree of heaven plantation experiment in eastern Austria. Aust J For Sci 132:241–256
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Qin H, Yang G, Provan J, Liu J, Gao L (2017) Using MiddRAD-seq data to develop polymorphic microsatellite markers for an endangered yew species. Plant Divers 39(5):294–299
Selkoe KA, Toonen RJ (2006) Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol Lett 9(5):615–629
Shokralla S, Porter TM, Gibson JF, Dobosz R, Janzen DH, Hallwachs W, Golding B, Hajibabaei M (2015) Massively parallel multiplex DNA sequencing for specimen identification using an Illumina MiSeq platform. Sci Rep 5:9687
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35(suppl_2):W71–W74
Vartia S, Villanueva-Cañas JL, Finarelli J, Farrell ED, Collins PC, Hughes GM, Carlsson JE, Gauthier DT, McGinnity P, Cross TF, FitzGerald RD, Mirimi L, Crispie F, Cotter PD, Carlsson J (2016) A novel method of microsatellite genotyping-by-sequencing using individual combinatorial barcoding. R Soc Open Sci 3(1):150565
Wang J (2016) Individual identification from genetic marker data: developments and accuracy comparisons of methods. Mol Ecol Resour 16(1):163–175
Weeks DE, Conley YP, Ferrell RE, Mah TS, Gorin MB (2002) A tale of two genotypes: consistency between two high-throughput genotyping centers. Genome Res 12(3):430–435
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370
Winter S (2018) Workflow for microsatellite genotyping from Illumina amplicon sequencing for non-model mammal species. Master thesis, FH Campus Vienna
Zhan L, Paterson IG, Fraser BA, Watson B, Bradbury IR, Nadukkalam Ravindran P, Reznick D, Beiko RG, Bentzen P (2017) MEGASAT: automated inference of microsatellite genotypes from sequence data. Mol Ecol Resour 17(2):247–256
Zhang J, Kobert K, Flouri T, Stamatakis A (2013) PEAR: a fast and accurate Illumina paired-end reAd mergeR. Bioinformatics 30(5):614–620
Zhao H, Yang L, Peng Z, Sun H, Yue X, Lou Y, Dong L, Wang L, Gao Z (2015) Developing genome-wide microsatellite markers of bamboo and their applications on molecular marker assisted taxonomy for accessions in the genus Phyllostachys. Sci Rep 5:80
Acknowledgments
Open access funding provided by University of Natural Resources and Life Sciences Vienna (BOKU). We greatly acknowledge the skilled technical assistance of Eva Dornstauder-Schrammel and Renata Milčevičova and would like to thank Benno Eberhard and Eugen Zimm for their assistance during field collections. We further thank all the forest companies and their representatives for their support in providing the forest stands and information about the history of Ailanthus in the three study stands. Finally, we are grateful to the anonymous reviewers for their careful reading of the manuscript and their useful comments that helped us improve the quality of the paper.
Funding
This work received support from the foundation “120 Years of the University of Natural Resources.”
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Data archiving statement
Genotypic data used for this study are available at Dryad: doi:https://doi.org/10.5061/dryad.873b2j2. All sequences are available in NCBI. A list of accession numbers is provided in Online Resource 4.
Additional information
Communicated by W.-W. Guo
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Neophytou, C., Torutaeva, E., Winter, S. et al. Analysis of microsatellite loci in tree of heaven (Ailanthus altissima (Mill.) Swingle) using SSR-GBS. Tree Genetics & Genomes 14, 82 (2018). https://doi.org/10.1007/s11295-018-1295-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-018-1295-4