Abstract
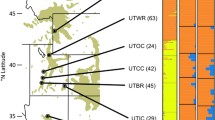
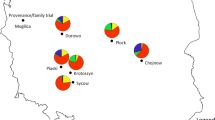
Forest trees dominate many Alpine landscapes that are currently exposed to changing climate. Norway spruce is one of the most important conifer species of the Italian Alps, and natural populations are found across steep environmental gradients with large differences in temperature and moisture availability. This study seeks to determine and quantify patterns of genetic diversity in natural populations toward understanding adaptive responses to changing climate. Across the Italian species range, 24 natural stands were sampled with a major focus on the Eastern Italian Alps. Sampled trees were genotyped for 384 selected single nucleotide polymorphisms (SNPs) from 285 genes. A wide array of potential candidate genes was tested for correlation with climatic parameters. To minimize false-positive association between genotype and climate, population structure was investigated. Pairwise F ST estimates between sampled populations ranged between 0.000 and 0.075, with the highest values involving the two disjoint populations, Valdieri, on the western Italian Alps, and Campolino, the most southern population on the Apennines. Despite considerable genetic admixture among populations, both Bayesian and multivariate approach identified four genetic clusters. Selection scans revealed five F ST outliers, and the environmental association analysis detected ten SNPs associated to one or more climatic variables. Overall, 13 potentially adaptive loci were identified, three of which have been reported in a previous study on the same species conducted on a broader geographical scale. In our study, precipitation, more than temperature, was often associated with genotype; therefore, it appears as the most important environmental variable associated with the high sensitivity of Norway spruce to soil water supply. These findings provide relevant information for understanding and quantifying climate change effects on this species and its ability to genetically adapt.



Similar content being viewed by others
References
Acheré V, Favre JM, Besnard G, Jeandroz S (2005) Genomic organization of molecular differentiation in Norway spruce (Picea abies). Mol Ecol 14:3191–3201
Albrechtsen A, Nielsen FC, Nielsen R (2010) Ascertainment biases in SNP chips affect measures of population divergence. Mol Biol Evol 27:2534–2547
Allen CD, Breshears DD (1998) Drought-induced shift of a forest-woodland ecotone: rapid landscape response to climate variation. Proc Natl Acad Sci U S A 95:14839–14842
Bar M, Ori N (2015) Compound leaf development in model plant species. Curr Opin Plant Biol 23:61–69
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier (France)
Beniston M, Diaz HF, Bradley RS (1997) Climatic change at high elevation sites: an overview. Climate Change 36:233–251
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57:289–300
Bergmann F, Ruetz W (1991) Isozyme genetic variation and heterozygosity in random tree samples and selected orchard clones from the same Norway spruce populations. For Ecol Manag 46:39–47
Berry J, Bjorkman O (1980) Photosynthetic response and adaptation to temperature in higher plants. Annu Rev Plant Physiol 31:491–543
Borghetti M, Giannini R, Menozzi P (1988) Geographic variation in cones of Norway spruce (Picea abies (L.) Karst.). Silvae Genet 37:178–184
Bragg JG, Supple MA, Andrew RL, Borevitz JO (2015) Genomic variation across landscapes: insights and applications. New Phytol 207:953–957
Canaani O, Havaux M, Malkin S (1986) Hydroxylamine, hydrazine and methylamine donate electrons to the photooxidizing side of photosystem II in leaves inhibited in oxygen evolution due to water stress. Biochim Biophys Acta BBA - Bioenerg 851:151–155
Chen J, Källman T, Ma X, Gyllenstrand N, Zaina G, Morgante M, Bousquet J, Eckert A, Wegrzyn J, Neale D, Lagercrantz U, Lascoux M (2012) Disentangling the roles of history and local selection in shaping clinal variation of allele frequencies and gene expression in Norway spruce (Picea abies). Genetics 191:865–881
Coop G, Witonsky D, Rienzo AD, Pritchard JK (2010) Using environmental correlations to identify loci underlying local adaptation. Genetics 185:1411–1423
De Mita S, Thuillet AC, Gay L, Ahmadi N, Manel S, Ronfort J, Vigouroux Y (2013) Detecting selection along environmental gradients: analysis of eight methods and their effectiveness for outbreeding and selfing populations. Mol Ecol 22:1383–1399
de Villemereuil P, Frichot E, Bazin E, François O, Gaggiotti OE (2014) Genome scan methods against more complex models: when and how much should we trust them? Mol Ecol 23:2006–2019
Ditmarová L, Kurjak D, Palmroth S, Kmet J, Střelcová K (2010) Physiological responses of Norway spruce (Picea abies) seedlings to drought stress. Tree Physiol 30:205–213
Earl DA, vonHoldt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Eckert AJ, van Heerwaarden J, Wegrzyn JL, Nelson CD, Ross-Ibarra J, Gonzalez-Martinez SC, Neale DB (2010) Patterns of population structure and environmental associations to aridity across the range of loblolly pine (Pinus taeda L., Pinaceae). Genetics 185:969–982
Endler JA (1986) Natural selection in the wild. Princeton University Press
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Hofer T, Foll M (2009) Detecting loci under selection in a hierarchically structured population. Heredity 103:285–298
Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303:1831–1838
Foll M, Gaggiotti O (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993
Giannini R, Morgante M, Vendramin GG (1991) Allozyme variation in Italian populations of Picea abies (L.) Karst. Silvae Genet 40:160–166
Ginzburg LR, Jensen CXJ (2004) Rules of thumb for judging ecological theories. Trends Ecol Evol 19:121–126
Ginzburg LR, Jensen CXJ, Yule JV (2007) Aiming at “unreasonable effectiveness of mathematics” at ecological theory. Ecol Model 207:356–362
Günther T, Coop G (2013) Robust identification of local adaptation from allele frequencies. Genetics 195:205-220
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New For 6:95–124
Hancock AM, Witonsky DB, Gordon AS, Eshel G, Pritchard JK, Coop G, Di Rienzo A (2008) Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet 4:e32
Haylock MR, Hofstra N, Klein Tank AMG, Klok EJ, Jones PD, New M (2008) A European daily high-resolution gridded data set of surface temperature and precipitation for 1950–2006. J Geophys Res-Atmos 113:D20119
Heuertz M, De Paoli E, Källman T, Larsson H, Jurman I, Morgante M, Lascoux M, Gyllenstrand N (2006) Multilocus patterns of nucleotide diversity, linkage disequilibrium and demographic history of Norway spruce [Picea abies (L.) Karst]. Genetics 174:2095–2105
Hickling R, Roy DB, Hill JK, Fox R, Thomas CD (2006) The distributions of a wide range of taxonomic groups are expanding polewards. Glob Chang Biol 12:450–455
Hortal J, Garcia-Pereira P, García-Barros E (2004) Butterfly species richness in mainland Portugal: predictive models of geographic distribution patterns. Ecography 27:68–82
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Initiative AG (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
IPCC (2014) Summary for policymakers. In: Edenhofer O, Pichs-Madruga R, Sokona Y, Farahani E, Kadner S, Seyboth K, Adler A, Baum I, Brunner S, Eickemeier P, Kriemann B, Savolainen J, Schlömer S, von Stechow C, Zwickel T, Minx JC (eds) Climate Change 2014: mitigation of climate change. Contribution of working group III to the fifth assessment report of the Intergovernmental Panel on Climate change. Cambridge University Press, Cambridge, United Kingdom and New York, NY, USA
Jaillon O, Aury J-M, Noel B et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Joost S, Vuilleumier S, Jensen JD, Schoville S, Leempoel K, Stucki S, Widmer I, Melodelima C, Rolland J, Manel S (2013) Uncovering the genetic basis of adaptive change: on the intersection of landscape genomics and theoretical population genetics. Mol Ecol 22:3659–3665
Karlsson PE, Medin EL, Wallin G, Selldén G, Skärby L (1997) Effects of ozone and drought stress on the physiology and growth of two clones of Norway spruce (Picea abies). New Phytol 136:265–275
Kass RE, Raftery AE (1995) Bayes factors. J Am Stat Assoc 90:773–795
Keller SR, Levsen N, Olson MS, Tiffin P (2012) Local adaptation in the flowering-time gene network of balsam poplar, Populus balsamifera L. Mol Biol Evol 29:3143–3152
Lagercrantz U, Ryman N (1990) Genetic structure of Norway spruce (Picea abies): concordance of morphological and allozymic variation. Evolution 44:38
Le Corre V, Kremer A (2012) The genetic differentiation at quantitative trait loci under local adaptation. Mol Ecol 21:1548–1566
Lehsten D, Dullinger S, Hulber K, Schurgers G, Cheddadi R, Laborde H, Lehsten V, Francois L, Drury M, Sykes MT (2014) Modelling the Holocene migrational dynamics of Fagus sylvatica L. and Picea abies (L.) H. Karst. Glob Ecol Biogeogr 23:658–668
Leimu R, Vergeer P, Angeloni F, Ouborg NJ (2010) Habitat fragmentation, climate change, and inbreeding in plants. Ann N Y Acad Sci 1195:84–98
Li Y, Stocks M, Hemmilä S, Källman T, Zhu H, Zhou Y, Chen J, Liu L, Lascoux K (2010) Demographic histories of four spruce (Picea) species of the Qinghai-Tibetan Plateau and neighbouring areas inferred from multiple nuclear loci. Mol Biol Evol 27:1001–1014
Lotterhos KE, Whitlock MC (2014) Evaluation of demographic history and neutral parameterization on the performance of F ST outlier tests. Mol Ecol 23:2178–2192
Lu C, Zhang J (1999) Effects of water stress on photosystem II photochemistry and its thermostability in wheat plants. J Exp Bot 50:1199–1206
Margules CR, Nicholls AO, Austin MP (1987) Diversity of Eucalyptus species predicted by a multi-variable environment gradient. Oecologia 71:229–232
Meloni M, Perini D, Binelli G (2007) The distribution of genetic variation in Norway spruce (Picea abies Karst.) populations in the western Alps. J Biogeogr 34:929–938
Metz M, Rocchini D, Neteler M (2014) Surface temperatures at the continental scale: tracking changes with remote sensing at unprecedented detail. Remote Sens 6:3822–3840
Mosca E, Eckert AJ, Di Pierro EA et al (2012) The geographical and environmental determinants of genetic diversity for four alpine conifers of the European Alps. Mol Ecol 21:5530–5545
Mosca E, González-Martínez SC, Neale DB (2014) Environmental versus geographical determinants of genetic structure in two subalpine conifers. New Phytol 201:180–192
Neale DB, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12:111–122
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Neteler M (2010) Estimating daily land surface temperatures in mountainous environments by reconstructed MODIS LST data. Remote Sens 2:333–351
Neteler M, Bowman MH, Landa M, Metz M (2012) GRASS GIS: a multi-purpose open source GIS. Environ Model Softw 31:124–130
Nystedt B, Street NR, Wetterbom A et al (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584
Palmer MW, McGlinn DJ, Fridley JD (2008) Artifacts and artifictions in biodiversity research. Folia Geobot 43:245–257
Pardo-Diaz C, Salazar C, Jiggins CD (2015) Towards the identification of the loci of adaptive evolution. Methods Ecol Evol 6:445–464
Partanen J, Koski V, Hänninen H (1998) Effects of photoperiod and temperature on the timing of bud burst in Norway spruce (Picea abies). Tree Physiol 18:811–816
Pawson T (1995) Protein modules and signalling networks. Nature 373:573–580
Peñuelas J, Boada M (2003) A global change-induced biome shift in the Montseny mountains (NE Spain). Glob Chang Biol 9:131–140
Piotti A, Leonardi S, Piovani P, Scalfi M, Menozzi P (2009) Spruce colonization at treeline: where do those seeds come from? Heredity 103:136–145
Prentice IC, Cramer W, Harrison SP, Leemans R, Monserud RA, Solomon AM (1992) A global biome model based on plant physiology and dominance, soil properties and climate. J Biogeogr 19:117
Pritchard JK, Di Rienzo A (2010) Adaptation–not by sweeps alone. Nature Rev Gen 11: 665-667.
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945
Prunier J, Laroche J, Beaulieu J, Bousquet J (2011) Scanning the genome for gene SNPs related to climate adaptation and estimating selection at the molecular level in boreal black spruce. Mol Ecol 20:1702–1716
Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17:369–381
Rellstab C, Gugerli F, Eckert AJ, Hancock AM, Holderegger R (2015) A practical guide to environmental association analysis in landscape genomics. Mol Ecol 24:4348–4370
Rennenberg H, Loreto F, Polle A, Brilli F, Fares S, Beniwal R, Gessler A (2006) Physiological responses of forest trees to heat and drought. Plant Biol 8:556–571
Rook DA (1969) The influence of growing temperature on photosynthesis and respiration of Pinus radiata seedlings. N Z J Bot 7:43–55
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Scalfi M, Mosca E, Di Pierro EA, Troggio M, Vendramin GG, Sperisen C, La Porta N, Neale DB (2014) Micro- and macro-geographic scale effect on the molecular imprint of selection and adaptation in Norway spruce. PLoS ONE 9
Schmidt-Vogt H (1977) Die Fichte. Verlag Paul Parey, Hamburg, Germany
Schuster R, Oberhuber W (2013) Age-dependent climate–growth relationships and regeneration of Picea abies in a drought-prone mixed-coniferous forest in the Alps. Can J For Res 43:609–618
Scotti I, Vendramin GG, Matteotti LS, Scarponi C, Sari-Gorla M, Binelli G (2000) Postglacial recolonization routes for Picea abies K. in Italy as suggested by the analysis of sequence-characterized amplified region (SCAR) markers. Mol Ecol 9:699–708
Seppä H, Schurgers G, Miller PA, Bjune AE, Giesecke T, Kühl N, Renssen H, Salonen JS (2015) Trees tracking a warmer climate: the Holocene range shift of hazel (Corylus avellana) in northern Europe. The Holocene 25:53–63
Sork VL, Aitken SN, Dyer RJ, Eckert AJ, Legendre P, Neale DB (2013) Putting the landscape into the genomics of trees: approaches for understanding local adaptation and population responses to changing climate. Tree Genet Genome 9:901–911
Sutinen R, Teirilä A, Pänttäjä M, Sutinen M-L (2002) Distribution and diversity of tree species with respect to soil electrical characteristics in Finnish Lapland. Can J For Res 32:1158–1170
Tabacchi G, De Natale F, Floris A, Gasparini P, Scrinzi G, Tosi V (2005) Surface estimation 2005—part one. INFC—National Inventory of Forests and Forest Carbon Sinks. MiPAF-State Forestry-General Inspectorate; CRA-ISAFA, Trento. 413 pp. [Italian]
Theurillat J-P, Guisan A (2001) potential impact of climate change on vegetation in the European Alps: a review. Clim Chang 50:77–109
Toivonen P, Vidaver W (1988) Variable chlorophyll a fluorescence and CO2 uptake in water-stressed White spruce seedlings. Plant Physiol 86:744–748
Tollefsrud MM, Kissling R, Gugerli F, Johnsen O, Skroppa T, Cheddadi R, van der Knaap WO, Latałowa M, Terhurne-Berson R, Litt T, Geburek T, Brochmann C, Sperisen (2008) Genetic consequences of glacial survival and postglacial colonization in Norway spruce: combined analysis of mitochondrial DNA and fossil pollen. Mol Ecol 17:4134–4150
Tollefsrud MM, Sønstebø JH, Brochmann C, Johnsen Ø, Skrøppa T, Vendramin GG (2009) Combined analysis of nuclear and mitochondrial markers provide new insight into the genetic structure of North European Picea abies. Heredity 102:549–562
Tsumura Y, Uchiyama K, Moriguchi Y, Ueno S, Ujino-Ihara T (2012) Genome scanning for detecting adaptive genes along environmental gradients in the Japanese conifer, Cryptomeria japonica. Heredity 109:349–360
Velasco R, Zharkikh A, Affourtit J et al (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–839
Vogel JG, Bond-Lamberty BP, Schuur EAG, Gower ST, Mack MC, O’Connell KEB, Valentine DW, Ruess RW (2008) Carbon allocation in boreal black spruce forests across regions varying in soil temperature and precipitation. Global Chang Biol 14:1503–1516
Walther GR, Post E, Convey P, Menzel A, Parmesan C, Beebee TJ, Fromentin JM, Hoegh-Guldberg O, Bairlein F (2002) Ecological responses to recent climate change. Nature 416:389–395
Wegrzyn JL, Eckert AJ, Choi M, Lee JM, Stanton BJ, Sykes R, Davis MS, Tsai CJ, Neale DB (2010) Association genetics of traits controlling lignin and cellulose biosynthesis in black cottonwood (Populus trichocarpa, Salicaceae) secondary xylem. New Phytol 188:515–532
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Xu Q, Bateman A, Finn RD et al (2010) Bacterial pleckstrin homology domains: a prokaryotic origin for the PH domain. J Mol Biol 396:31–46
Yeaman S, Whitlock MC (2011) The genetic architecture of adaptation under migration–selection balance. Evolution 65:1897–1911
Acknowledgments
This work was supported by the Alpine Ecosystems in a Changing Environment: Biodiversity Sensitivity and Adaptive Potential (ACE-SAP) project c/o the Edmund Mach Foundation–San Michele all’Adige (TN) and partially funded by the Autonomous Province of Trento (Italy), with the regulation No. 23, June 12, 2008, of the University and Scientific Research Service.
The authors would wish to thank Alessandro Mancabelli, soil expert at FEM, Marco Pietrogiovanna of the Forest Service of the Autonomous Province of Bolzano, Raffaella Pettina of the Italian Forest Service, Paolo Camerano of IPLA-Turin, and Luca Bronzini and Maurizio Odasso of Panstudio Ass. A special thank to Antonella Agostini, Lucio Sottovia, Fabio Angeli, Giorgio Messina, Bruno Crosignani, Alessandro Wolinski, and Romano Masè of the Forest Services of the Provinces of Trento and Udine for their help in the sampling organization and David Blanco, Yuri Gori, Stefano Maffei, Marta Scalfi, and Daniele Sebastiani for their helpful support in sample collection. We would like to thank Randi Famula for DNA extraction and Ben Figueroa and Gabriel G. Rosa for bioinformatics support. A particular acknowledgment is for Jill L. Wegrzyn and John D. Liechty for their help in the genotyping design and data processing and Andrew J. Eckert for his comments on preliminary data analysis. We thank Luca Delucchi, Markus Metz, and Markus Neteler for providing GIS-derived climate data. The authors are also very thankful to the anonymous reviewers that provided important suggestions to improve the manuscript.
Data archiving statement
SNP sequence data is available at DRYAD digital repository with submission code doi:10.5061/dryad.n818s
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Gugerli
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
Supplementary figures from Fig. S1 to Fig. S6 (PDF 1.22 mb)
Online Resource 2
Supplementary tables from Table S1 to Table S7 (PDF 2.64 mb)
Rights and permissions
About this article
Cite this article
Di Pierro, E.A., Mosca, E., Rocchini, D. et al. Climate-related adaptive genetic variation and population structure in natural stands of Norway spruce in the South-Eastern Alps. Tree Genetics & Genomes 12, 16 (2016). https://doi.org/10.1007/s11295-016-0972-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-0972-4