Abstract
Twenty-nine provenances of teak (Tectona grandis Linn. f.) representing the full natural distribution range of the species were genotyped with microsatellite DNA markers to analyse genetic diversity and population genetic structure. Provenances originating from the semi-moist east coast of India had the highest genetic diversity while provenances from Laos showed the lowest. In the eastern part of the natural distribution area, comprising Myanmar, Thailand and Laos, there was a strong clinal decrease in genetic diversity the further east the provenance was located. Overall, the pattern of genetic diversity supports the hypothesis that teak has its centre of origin in India, from where it spread eastwards. The analysis of molecular variance (AMOVA) gave an overall highly significant F st value of 0.227—population pairwise F st values were in the range 0.01–0.48. Applying the G″st differentiation parameter, the estimated overall differentiation was 0.632, implying a strong genetic structure among populations. A neighbour-joining (NJ) tree, using the pairwise population matrix of G″st values as input, contained three distinct groups: (1) the eight provenances from Thailand and Laos, (2) the Indian provenances from the dry interior and the moist west coast and (3) the provenances from northern Myanmar. The provenances from southern Myanmar were placed close to the root of the tree together with the three provenances from the semi-moist east coast of India. A Bayesian cluster analysis using the STRUCTURE software gave very similar results, with three main clusters, each containing two sub-clusters, while Bayesian cluster analysis in the Geneland software, exploiting the spatial coordinates of the provenances, resulted in five clusters in accordance with the former results. The implications of the findings for conservation and use of genetic resources of the species are discussed.
Similar content being viewed by others
Introduction
Teak (Tectona grandis Linn. f.) is a deciduous tree belonging to the Lamiaceae family (The International Plant Names Index 2014) and one of the most important timber species of the world. Its wood is renowned for qualities such as resistance to weather and other causes of decay, as well as its elasticity and solid fibre, both of which facilitate woodworking. Furthermore, the wood possesses a high dimensional stability under changing temperature and humidity (Tsoumis 1991).
Teak has a large—to some extent geographically dispersed—natural distribution area encompassing parts of India, Myanmar, Thailand and Laos (Kaosa-ard 1981). It also grows widely on Java, where presumably it was introduced between the beginning of the fourteenth and sixteenth centuries (Verhaegen et al. 2010). The species grows naturally in a wide range of environmental conditions from dry places with only 500 mm annual rain up to areas with 5000 mm—but optimum conditions are in areas having between 1200 and 2500 mm rainfall annually (Kaosa-ard 1981). Teak is a light-demanding (Soerianegara and Lemmens 1994), mainly insect-pollinated and outcrossing species (Bryndum and Hedegart 1969; Hedegart 1973; Kjær and Suangtho 1995; Kertadikara and Prat 1995a; Tangmitcharoen and Owens 1997).
Due to the factors described above—the valuable wood and its pioneer species traits, which make it suitable for plantations—teak has become one of the most important tree species for high-grade wood plantations in the tropics (Kollert and Cherubini 2012). This is the case across Southeast Asia, but during the last 30–40 years, teak has also become an important tree species for plantations outside its natural distribution area. The potential value of teak for exotic plantations was recognised early in many countries; seed was therefore transferred to Africa and Central America over 100 years ago. Teak has been reported to have been introduced successfully to Nigeria, the West Indies, Trinidad and Papua New Guinea before 1900 and to Tanzania, Togo and Ghana just after the turn of the century, while introductions to Cote d’Ivoire, Sudan and a number of additional Central and South American countries are reported during the early decades of the twentieth century (see Verhaegen et al. 2010; Koskela et al. 2010 for references). Most early introductions were on a pilot basis, while large-scale planting was only initiated more recently. Seeds from the early pilot introductions have often formed the basis for the later scaling up of plantation forestry, and this is likely to have led to the formation of various landraces. Domestication and tree improvement were initiated in Thailand, India and Indonesia in the 1950s and 1960s and was followed up based on comprehensive selections of superior plus-trees in these countries (Keiding et al. 1986; Kjær and Foster 1996; Kaosa-ard et al. 1998). Since then, improvement activities have been initiated in many countries (Kjær et al. 2000), and genetic parameters estimated based on progeny and clonal trials in general predict good response to breeding (Mandal and Chawhan 2005; Callister and Collins 2008; Narayanan et al. 2009; Callister 2013).
Despite the huge economic value, there have been relatively few genetic studies of teak, and an exhaustive study comprising populations from the whole natural distribution area has never been conducted. Both in relation to conservation of genetic resources of the species and especially to the origin of the various landraces around the world, such a study could be beneficial.
Many of the breeding and domestication activities mentioned above are based on the local landraces, where the geographic and therefore genetic origin is unknown. Historical records have shed light on the likely origin of some major sources—e.g. Keogh (1981) made a valuable effort to document introduction to Central America, while Wood (1967) looked into the origin of East African landraces. Genetic approaches have also been tested: Kjær and Siegismund (1996) used allozyme markers to study landraces from Tanzania and Nicaragua, Fofana et al. (2008) studied the likely origin of West African landraces based on simple sequence repeat (SSR) markers on trees sampled in African provenance trials and Verhaegen et al. (2010) used the same markers to decide from where the teak growing in Africa and Indonesia has been introduced. Additional studies primarily focused on the genetic diversity of populations within the native range have been conducted using genetic markers (Kertadikara and Prat 1995b; Kjær et al. 1996; Changtragoon and Szmidt 2000; Changtragoon 2001; Nicodemus et al. 2005; Shrestha et al. 2005; Fofana et al. 2009; Sreekanth et al. 2012). However, although representing valuable contributions, none of the studies mentioned here have covered the whole distribution area of teak; rather, they have often been based on a limited number of populations and/or trees. Most importantly, they all lack samples from populations in Myanmar, which constitutes a significant part of the natural range with an estimated area of 16.5 million ha of natural teak, out of a total of 27.9 million ha (Gyi and Tint 1998).
The objective of the present study is therefore to make a comprehensive study of the genetic diversity and differentiation among provenances of teak over its whole natural distribution range using neutral DNA markers and based on sample sizes that allow range-wide comparison of diversity measures.
Materials and methods
Plant material
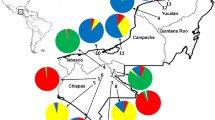
Provenances from the natural distribution area of teak were collected from two sites included in the international series of teak provenance trials described by Keiding et al. (1986). Seeds for these provenance trials were collected in 1971–1972. The regulations for provenance sampling concerning minimum number of trees and distance between parent trees, as prescribed by IUFRO (Lines 1965), were followed as far as possible. In the circular letter to the partners involved in the collection, it was emphasised that ‘it is important that the samples are representative of the stands from which they are taken, which means that seeds should be collected evenly from the whole of the area and not from a few trees’ (Danish/FAO Forest Tree Seed 1971). Seed from individual trees were mixed (Keiding et al. 1986). All collections were documented on seed collection sheets kept in copies at the Danida Forest Seed Centre (now Research Group of Tropical Trees and Landscapes, University of Copenhagen, Denmark). Samples from 17 provenances were collected from the field trial in Pha Nok Kao (Khon Kaen province) in Thailand (trial no. 038). For a further description of this trial and plant material, see Keiding et al. (1986) and Kjær et al. (1996). Samples from three Indian provenances not included in trial no. 038 were collected from the field trial in Huey Som Poi (Lampang province), also in Thailand (trial no. 001 according to Keiding et al. 1986). Samples from one additional Indian provenance collected in a provenance trial in Longuza, Tanzania (Provenance TAN_Coi; Persson 1971) were kindly shared by Tanzania Forest Research Institute and included in the present study. Sampling of trees in the abovementioned provenance trials for the present study was done as random sampling of remaining trees up to 31 individuals (i.e. without deliberate selection). Lastly, eight populations were sampled in situ by collecting leaves from adult trees (i.e. not from regeneration) from the natural distribution area in Myanmar. To minimise the risk of sampling related trees, the minimum distance between consecutive sampled adult trees was approximately 100 m (Minn 2012). Four of the populations sampled were in the northern region of Myanmar, while the remaining four populations were in the southern region of Myanmar, and all eight populations are presumed unlogged. All samples were taken as leaf material and stored in plastic bags with silica gel until DNA extraction took place. Up to 31 (N) trees were sampled from each provenance (average 24.9, median 25). For further details about the 29 provenances and their origins, see Table 1 and Fig. 1.
Laboratory work
DNA was extracted from the leaf tissue with one of three different extraction methods: (1) a modified version of the CTAB method of Doyle and Doyle (1990), (2) the QIAGEN® (Germany) DNeasy Plant Mini Kit or (3) the QIAGEN® DNeasy 96 Plant Kit.
All individuals were genotyped with 6 microsatellites out of 15 developed by Verhaegen et al. (2005). The six microsatellites were chosen according to three criteria: high polymorphism, small difference between observed and expected heterozygosity (H o and H e) and whether amplification products could be combined in one single multiplex PCR. The six analysed microsatellites were CIRAD1TeakA06, CIRAD1TeakB03, CIRAD2TeakB07, CIRAD2TeakC03, CIRAD3TeakA11 and CIRAD3TeakF01 (Verhaegen et al. 2005).
Genotyping of microsatellites took place in 10 μl PCRs using the QIAGEN® Multiplex Kit (catalogue no. 206143). PCR conditions followed the given standard multiplex PCR protocol: 1× Multiplex master mix (providing a final concentration of 3 mM MgCl2), 0.2 μM of each primer, and around 20 ng of DNA sample with water added to make the final reaction volume. Amplifications were carried out in a Bio-Rad thermal cycler (model C1000) with the following thermal profile: 15 min of denaturation at 95 °C, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 90 s and extension at 72 °C for 60 s, with a final extension step at 60 °C for 30 min.
Fragment sizes were determined on an ABI 3130XL genetic analyser and analysed with the GeneMapper software version 4.0 (Applied Biosystems).
Genetic data analysis
Genetic diversity in the sampled provenances was estimated for each locus using the following parameters: observed heterozygosity (H o), expected heterozygosity (H e), number of observed alleles (Na), effective number of alleles (Ne) and allelic richness calculated via rarefaction (Na(rar)). The first measures were calculated in GenAlEx ver. 6.5 (Peakall and Smouse 2006, 2012), and allelic richness was calculated in the HP-Rare 1.1 software (Kalinowski 2005). While Na is the raw number of observed alleles, Ne estimates the number of equally frequent alleles it would take to achieve a given level of genetic diversity in a panmictic population, a number which will usually be less than the actual number (Kimura and Crow 1964). Na(rar) is the number of observed alleles in a provenance, corrected for sample size through rarefaction, since larger samples are expected to have more alleles than smaller samples (Kalinowski 2004). The fixation index, F (Hartl and Clark 1997), was also calculated in GenAlEx.
To explore geographic patterns in genetic diversity, based on the initial observations, we plotted the allelic richness Na(rar) of each provenance against its position on a west-east transect starting at the most western provenance (prov. 3013 at longitude 74° 41). The distance of each provenance from longitude 74° 41 was calculated in kilometres—this was done at the website of the USA National Hurricane Center (http://www.nhc.noaa.gov/gccalc.shtml). The latitude of prov. 3013 was used to calculate the position of all provenances on this transect.
The genetic structure of the sampled populations was analysed with an analysis of molecular variance (AMOVA; Excoffier et al. 1992) performed in Arlequin ver. 3.5.1.3 (Excoffier et al. 2005), including calculation of pairwise F st values between populations. The ability of F st to truly describe the genetic differentiation between populations is the subject of continuing discussion—especially in relation to highly polymorphic markers such as microsatellites (Hedrick 2005; Jost 2008). This has resulted in the development of several alternative estimators for population differentiation; for this study, two of these—Jost’s D (Jost 2008) and G″st (Meirmans and Hedrick 2011)—were calculated and tested with G statistics in GenAlEx. To test for isolation by distance, we performed Mantel tests (Mantel 1967) between a matrix of pairwise geographic distances between populations and the matrices of pairwise F st values and G″st values, respectively. The geographic distance matrix was calculated in the Geographic Distance Matrix Generator software (Ersts), while the Mantel tests were performed in GenAlEx with 9999 permutations.
We consider the matrix of pairwise differentiation estimators between populations as the most appropriate illustration of the inter-population structure. However, although populations, unlike species, cannot be expected to have a tree-like phylogeny which can be illustrated with bifurcating trees, such trees are often used to depict the genetic relationships between populations (Kalinowski 2009). We therefore constructed a tree using the neighbour-joining (NJ) method (Saitou and Nei 1987), with the pairwise population differentiation matrices as input. This was done in the TreeFit software (Kalinowski 2009), which also can calculate the proportion of variance in the genetic distance matrix that is explained by the tree (R 2). The resulting tree was depicted in the graphical viewer FigTree ver. 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/).
As an alternative approach to describing the genetic structure of teak, Bayesian cluster analyses were conducted using the STRUCTURE software (version 2.3) (Pritchard et al. 2000) and the Geneland software (version 4.0.3) (Guillot et al. 2005a, b). STRUCTURE uses a Bayesian iterative algorithm to probabilistically assign individuals (in this instance trees) into K clusters in such a way that the genotyped loci are in Hardy-Weinberg equilibrium and linkage equilibrium within the clusters. Ten clustering runs were made for each K from 1 to 8, each with a burn-in time and run length of 100,000. Because of the strong genetic structure observed in the previous analyses, the ‘no admixture ancestry’ model was chosen in combination with the assumption of independent allele frequencies among populations. To infer the true number of clusters (K), we used the delta K method developed by Evanno et al. (2005) and the STRUCTURE HARVESTER software (Earl and VonHoldt 2012) to implement the method. Since the clustering in STRUCTURE involves stochastic simulation, replicate cluster analysis of the same data (i.e. our 10 replication runs for each K value) may produce different solutions due either to ‘label switching’ or ‘genuine multimodality’ (Jakobsson and Rosenberg 2007). We therefore used the CLUMPP software (Jakobsson and Rosenberg 2007) to make an alignment of the outcome of the 10 replicate cluster analyses with the identified optimal number of clusters (K). The DISTRUCT software (Rosenberg 2004) was used to illustrate visually the outcome of this alignment.
Geneland uses an admixture model where the likelihood is similar to the admixture model in STRUCTURE but where it is possible to include the geographic positions as an additional parameter in the analysis. The latter makes it possible to identify geographic boundaries between the identified clusters, and the spatial positions of the analysed provenances were therefore included. Ten runs, each including 200,000 Markov chain Monte Carlo (MCMC) iterations with a thinning of 200, were made, based on the uncorrelated frequency model with an uncertainty of 5 km attached to the spatial coordinates, and where the unknown number of clusters (K) was allowed to vary between 1 and 10. Excluding the first 200 values out of the 1000 saved iterations as a burn-in, MCMC convergence was assessed by comparing the number of clusters across replicate runs, with the mean posterior probability used as a criterion to choose the best run.
Results
Genetic diversity
The estimated genetic diversity in each of the 29 provenances can be seen in Table 2; means for the different regions are shown in Table 3. Clear differences among both provenances and regions were observed for all diversity parameters. The observed number of alleles (Na) and the allelic richness calculated via rarefaction (Na(rar)) showed a high correlation of 0.98 even though there is an up to threefold difference in sample sizes (minimum size N = 10, maximum N = 31). These two parameters can therefore be used interchangeably. The provenances originating from the semi-moist east coast of India (prov. 3033–3036) were most diverse. On average, these three provenances had 34 % more alleles (Na) than the overall mean across all 29 populations—although there was some variation within this region. At the opposite end of the spectrum, the two provenances from Laos (prov. 3055 and prov. 3057) clearly showed the lowest genetic diversity—although some of the Thai provenances also had diversity almost as low (e.g. prov. 3038 and prov. 3043). The two Laotian provenances had around 60 % fewer alleles than the overall mean of the study—the reduction in diversity was a little less pronounced for other diversity parameters such as Ne and heterozygosity. The two Laotian provenances were very similar in all their diversity estimates although they were taken from relatively disparate geographic locations (Fig. 1). The eight provenances from Myanmar showed a clear tendency to greater genetic diversity in the southern region—especially in terms of Na. The four provenances from northern Myanmar were intermediate with values very close to the overall mean for all parameters, while the values for southern Myanmar provenances were a little higher. Most of the group of Thai provenances, as mentioned above, had low genetic diversity, while a provenance such as prov. 3042 was similar to the overall mean for the study; the average Na in the populations from Thailand was more than 35 % lower than the overall mean.
The geographic distribution of genetic diversity showed decreasing diversity from west to east in the distribution area (see Fig. 1 and Table 2). Three plots of allelic richness (Na(rar)) against a position on a west-east transect were made: one for all 29 provenances, one for the Indian provenances and one for the remaining provenances (Fig. 2). As seen in Fig. 2a, when allelic richness for all 29 provenances was plotted on the same figure, there was clear difference between the observed pattern in the Indian part of the distribution area compared to the region including Myanmar, Thailand and Laos. In Fig. 2b, c, the allelic richness for these two regions was therefore plotted separately. For the Indian region, there was almost no relationship between Na(rar) and the position on the west-east transect, whilst for the eastern region of the distribution area, there was a very strong cline with decreasing allelic richness towards the east. In fact, using a linear regression, 88 % of the variation in Na(rar) can be explained by position on the west-east transect (Fig. 2c)—giving a correlation of approximately 0.94.
Plots of the allelic richness Na(rar) for each provenance against position on a west-east transect starting at the most western provenance (prov. 3013 at longitude 74° 41). a Plot of Na(rar) against position on the west-east transect in kilometres for all 29 provenances. b Plot of Na(rar) against position on the west-east transect—Indian provenances only. c Plot of Na(rar) against position on the west-east transect—Myanmar, Thai and Laotian provenances only
The overall mean for the fixation index (F) was 0.036—but with large variation between provenances. In general, the provenances from Thailand had high fixation indices and prov. 3039 had the highest F value, 0.30, but also two provenances from West India (prov. 3019 and prov. 3020) had F values above 0.20. Several provenances had F values below zero, but all of them were of small magnitude.
Population differentiation
The AMOVA gave a highly significant overall F st value of 0.227 (among population variance), and population pairwise F st values were in the range 0.01–0.48 (Table 4—lower triangle).
The structure among the populations expressed through the pairwise F st values is illustrated in Fig. 3, which gives a more easily comprehensible overview of the pattern of genetic differentiation. The two Laotian provenances (prov. 3055 and prov. 3057) were highly differentiated from both the Indian and Myanmar provenances (Fig. 3)—almost all the pairwise F st values being close to 0.40 (Table 4). The F st values were below 0.20 only to some of the Thai provenances, and even the two Laotian provenances, which had very similar diversity estimates, showed substantial genetic differentiation with F st = 0.24 (Table 4 and Fig. 3).
Using the G″st differentiation parameter developed by Meirmans and Hedrick (2011), we estimated overall differentiation to be 0.632, almost three times the size of the overall F st value. The overall Jost’s D was estimated to be 0.526. Both G″st and Jost’s D were statistically highly significant in the corresponding G statistics test.
When analysing the pairwise values between provenances, G″st and Jost’s D showed a very strong correlation of 0.99, so these two measures basically captured the same genetic pattern, the only difference being in terms of the level of differentiation, with G″st indicating higher genetic differentiation. Because of the strong correlation, pairwise population values are shown only for G″st (Table 4—upper triangle)—with G″st values being in the range 0.02–0.94. The correlation between pairwise estimates of F st and G″st (values in the lower and upper triangles of Table 4) was somewhat lower, at 0.91.
The Mantel tests between pairwise geographic distances between populations and the matrices of pairwise F st values and G″st values, respectively, were highly significant with a p value of 0.0001 in both cases. The correlation coefficient between pairwise geographic distances and pairwise F st values was 0.49, while it was 0.64 with pairwise G″st values.
When using the pairwise population matrix of G″st values as input to make a NJ tree in the TreeFit software, the proportion of variance in the genetic distance matrix explained by the tree (R 2) was 0.84. The NJ tree (Fig. 4) depicted three distinct groups: (1) the eight provenances from Thailand and Laos cluster together in the upper part of the tree; (2) the Indian provenances from the dry interior and the moist west coast cluster together in the middle of the tree and (3) the provenances from northern Myanmar make a third distinct cluster at the bottom of the tree. The provenances from southern Myanmar are placed close to the root of the tree together with the three provenances from the semi-moist east coast of India (Fig. 4).
Neighbour-joining tree using the pairwise population matrix of G″st values as input (genetic distance matrix). Colour code: yellow = India—dry interior, dark green = India—moist west coast, magenta = India—semi-moist east coast, light green = Myanmar—northern region, cyan = Myanmar—southern region, blue = Thailand, red = Laos
The Bayesian cluster analysis with STRUCTURE showed that the most likely number of clusters (K) was 3, using the delta K method (Evanno et al. 2005). This result was achieved consistently even when the model or assumptions were changed. The three main clusters are depicted in Fig. 5. These are the output from the 10 STRUCTURE runs which were aligned in the CLUMPP software and illustrated in the DISTRUCT software. Cluster 1 (light green) consists of the Indian provenances from the dry interior and the moist west coast, cluster 2 (orange) encompasses the provenances from the semi-moist east coast of India plus the Myanmar provenances and cluster 3 (blue) comprises the Thai and Laotian provenances. Running a new AMOVA, including the three clusters as regions, the partition of molecular variance was as follows: 18.1 % among regions (equal to three clusters), 9.0 % among populations within regions and 72.9 % among individuals within populations.
Illustration of results from STRUCTURE analysis with K = 3 clusters, where the 10 replicate cluster analyses have been aligned in CLUMPP. Each of 29 populations (721 trees in total) is represented with a horizontal bar. Vertical height of the bar represents the number of trees in the population (range 10–31). For each population, an estimated cluster membership coefficient is shown, bar colours indicate the clusters: (1) light green = India—dry interior and moist west coast, (2) orange = India—semi-moist east coast and Myanmar, (3) blue = Thailand and Laos
To search for population structures at a lower level, a second round of analysis was performed with STRUCTURE for the provenances within each of the three main clusters. As the provenances within the main clusters are closer, both the previous model (‘no admixture ancestry’ model combined with the assumption of independent allele frequencies among populations) and the ‘admixture ancestry’ model combined with the assumption of allele frequencies being correlated among populations were tested. These models gave very similar results—here, we report the results from the latter only.
In all three cases, two sub-clusters were revealed within the main clusters using the delta K method. The six sub-clusters are depicted in Fig. 6. In Fig. 6a, cluster 1 from the first STRUCTURE analysis is shown with two sub-clusters, one consisting of the three provenances from the dry interior of India plus prov. 3013 which is located very close to prov. 3008. The other sub-cluster comprises the remaining six provenances from the moist west coast of India. Both cluster 1 and its sub-clusters are in line with the NJ tree in Fig. 4. The sub-clustering for cluster 2 is illustrated in Fig. 6b; the provenances from the east coast of India form a sub-cluster with the southern Myanmar provenances, whilst the northern Myanmar provenances form a different sub-cluster. Again, this result is in line with the NJ tree based on G″st values (Fig. 4). This is also the case for the last main cluster comprising the Thai and Laotian provenances (Fig. 6c), where one sub-cluster comprises the most western Thai provenances (provs. 3039, 3041 and 3042) and the remaining Thai provenances and the two Laotian provenances make up the other sub-cluster. The former sub-cluster is also separated slightly from the others in the main cluster on the NJ tree, with all three provenances of this sub-cluster being closer to the root of the tree (Fig. 4).
Using the spatial model in Geneland, assuming uncorrelated allele frequencies, the highest mean posterior density was obtained for K = 5 in all 10 replicate runs. A map of the estimated cluster membership for the five clusters is seen in Fig. 7. Besides the number of clusters being 5 and not 6 as found in STRUCTURE (including sub-clusters), the results are very similar among the two Bayesian clustering methods. Both agree that there is a north-west and a south Indian cluster and that provenance 3013 is around the border between these two—although assigned to the north-west (compare Figs. 6a and 7). They also agree on the existence of an Eastern Indian and southern Myanmar cluster and a distinct north Myanmar cluster (compare Figs. 6b and 7) and that prov. 3036 is just on the border between the north-west Indian cluster and the Eastern Indian and southern Myanmar cluster (Figs. 5 and 7). One discrepancy between the STRUCTURE and Geneland results is that the latter only identifies one cluster for the Thai and Laotian provenances and not two as in STRUCTURE (compare Figs. 6c and 7).
Discussion and conclusions
Despite the economic value of teak, the species is remarkably understudied scientifically—this includes studies of the genetic diversity and population structure of the species. The latter is particularly problematic because the provenance variation in both economically and ecologically important traits is huge (Keiding et al. 1986; Kjær et al.1995; Kjær et al. 1999; Kjær 2005; Pedersen et al. 2007; Monteuuis et al. 2011; Chaix et al. 2011).
The first studies of genetic diversity in teak in its natural distribution area with molecular markers were conducted with allozymes in the mid 1990s (Kertadikara and Prat 1995a, b; Kjær et al. 1996). Even with these markers, a higher genetic diversity was observed in Indian provenances than in Thai provenances (Kjær et al. 1996). Later studies using DNA-based markers such as RAPD (Nicodemus et al. 2005; Parthiban et al. 2005), AFLP (Shrestha et al. 2005; Sreekanth et al. 2012) and ISSR (Ansari et al. 2012) have also been performed—but these studies have mainly focused on Indian teak (Nicodemus et al. 2005; Parthiban et al. 2005; Ansari et al. 2012; Sreekanth et al. 2012) or sampled a very limited number of populations and individuals (Shrestha et al. 2005). Only the study by Fofana et al. (2009) can easily be compared to our study because there is an overlap both in SSR markers and in some of the provenances. Fofana et al. (2009) collected samples in the international provenance trials of teak established in Cote d’Ivoire and Ghana and part of the same seed collections which were the basis of the trials in Thailand used in the present study, resulting in an overlap of five provenances between the two studies. Given the limited sample size applied by Fofana et al. (2009) (166 genotyped individuals to sample the distribution area), it is interesting that their results in terms of general structure and diversity levels fit well with the findings in the present study as discussed below, and this in itself emphasises the presence of substantial and very distinct patterns of genetic diversity in teak. Fofana et al. (2009), however, only comprised 17 provenances with only 1 population from the Indian semi-moist east coast (prov. 3034) and none from Myanmar. Therefore, they had data from only 1 of the 11 provenances which we identify as forming cluster 2 (Fig. 5) in the present study.
We found very strong differences in genetic diversity across the distribution area—with the greatest diversity in the Indian provenances and the lowest in Laos—a three- to fourfold difference, which is in line with the findings of Fofana et al. (2009). In the first ever comparison of the genetic diversity of Myanmar teak with teak from the rest of the distribution area, we observed that Myanmar teak is not among the genetically most diverse but close to the global average. Thai provenances showed a lower genetic diversity although not as low as Laotian provenances. Generally, in the eastern region of the natural distribution area, a strong clinal decrease in genetic diversity (allelic richness) west to east was detected. These observations support the hypothesis that teak has its centre of origin in India, where the genetic diversity is highest, and spread eastwards from there. During this process, the founding populations may have experienced repeated founder effects (Holgate 1966) or periods with small population size, creating strong genetic drift and leading to low genetic diversity in the Laotian provenances on the margin of the distribution area. The previous connectivity between the Indian provenances and the remaining parts of the distribution area was shown in the analysis of population differentiation, where one cluster included provenances from both India and Myanmar. The highest diversity level was observed in the semi-moist east Indian teak populations, and this could point towards this area as the centre of origin within the Indian subcontinent. However, more detailed studies will be needed to clarify this because other areas with relative high levels of diversity were observed, and the variation in diversity level may reflect temporal differences in effective population numbers between areas, for example as a result of different climatic conditions in previous periods. The occasional high values for the fixation index F showed that there can be substantial inbreeding in the teak provenances. On the other hand, several provenances had negative F values, mostly of small magnitude, indicating that these negative estimates are mainly due to sampling error and do not reflect a real excess of heterozygotes. In some of the Myanmar provenances though, negative F values of larger magnitude and relatively small standard errors are less likely to be caused solely by sampling effects. In these cases, an excess of heterozygotes could be caused by negative assortative mating or selection for heterozygotes.
Population differentiation was strongly reflected by the overall F st value of 0.227. Thus, approximately 23 % of the genetic variation is distributed among populations, a result very similar to that found by Fofana et al. (2009). The high pairwise differentiation between some provenances—F st values up to 0.48 and G″st values up to 0.94—suggests that teak formed distinct geographic races, knowledge which can be used in sustainable utilisation of the species. The two different approaches of analysing population differentiation—pairwise estimates of F st/G″st values with subsequent NJ tree and Bayesian cluster analyses—gave very similar results, thereby providing confirmation of the findings.
Our cluster analysis in STRUCTURE, showing three main clusters, accords well with the traditional perception that teak has three main distribution areas—and the identification of two sub-clusters in each of those contributes to a more comprehensive understanding of the genetic structure of the species. The substantial genetic differentiation in terms of F st/G″st values is supported by observed phenotypic differences between provenances from these major clusters reported from the provenance field trials at several sites (Keiding et al. 1986; Kjær et al. 1995). The results stress the importance of developing gene conservation programmes that cover all parts of the gene pool. As mentioned earlier, Fofana et al. (2009) did only sample one of the provenances from our cluster 2 (STRUCTURE), but they found that the 10 individuals they genotyped from prov. 3034 were strongly differentiated from all other populations and grouped into a distinct cluster. On the more detailed level, our analysis revealed differentiation between the most western Thai provenances and the eastern Thai and Laotian provenances. This separation was already observed in the analysis of growth parameters and branching habits in the Thai provenance trial 038 (Kjær et al. 1996) and therefore implemented in the guidelines on gene conservation of teak in Thailand (Graudal et al. 1999). Based on genecological zonation according to ecotypes, this west-east pattern was not predicted because climatic gradients in Northern Thailand are mainly north-south and Graudal et al. (1999) therefore speculated that the west-east genetic structure was the result of a barrier to gene flow formed by the north-south-heading mountain ranges, where teak only occurs naturally in the lowlands separated by the mountain ridges with no natural teak and limited option for transfer of the relatively large fruits or small pollinating insects. The pronounced clinal loss of genetic diversity along the west-east gradient from Myanmar to Laos (cf. Fig. 2c) is one of the intriguing results of the present study, and in line with Graudal et al. (1999), we hypothesise that the pattern could result from repeated bottlenecks and/or founder effects generated during species expansion across physiographic barriers in Northern Thailand. More detailed analysis of vegetation history will be required to support such a hypothesis, but from a conservation perspective, it stresses the importance of protecting the remaining native teak forests in Eastern Thailand.
In Myanmar, the study revealed an interesting separation between the northern and southern part of the distribution area (cf. Fig. 7). This prediction of substantial genetic differentiation is in line with findings from detailed provenance trials in Myanmar, where Lwin et al. (2010) found differentiation in growth and quality traits following the latitudinal gradient. The finding of a unique northern genetic cluster in Myanmar stresses the importance of a focused gene conservation effort in the area, as the results suggest that the northern populations are likely to represent a unique gene pool within teak. We hope that the genetic patterns within and between the major regions reported in the present study in similar ways can support local efforts to develop and refine effective gene conservation efforts of the species. Although marker-based studies cannot replace testing of differentiation in traits under selection in common gardens or field trials (Graudal et al. 1997), the present results do demonstrate the importance of supporting conservation efforts with marker-based studies.
Another conservation aspect of the present work is related to the genetic origin of global landraces that represent a genetic resource in many teak-growing countries outside the natural distribution area of the species. The results of the present study can serve as reference to test the origin of landraces and especially to what extent different landraces represent single or multiple introductions from various origins. A knowledge which we trust can support sustainable domestication of genetic resources of teak in several countries.
References
Ansari SA, Narayanan C, Wali SA, Kumar R, Shukla N, Rahangdale SK (2012) ISSR markers for analysis of molecular diversity and genetic structure of Indian teak (Tectona grandis L.f.) populations. Ann For Res 55:11–23
Bryndum K, Hedegart T (1969) Pollination of teak (Tectona grandis Linn. f.). Silvae Genet 18:77–80
Callister AN (2013) Genetic parameters and correlations between stem size, forking, and flowering in teak (Tectona grandis). Can J Forest Res 43:1145–1150
Callister AN, Collins SL (2008) Genetic parameter estimates in a clonally replicated progeny test of teak (Tectona grandis Linn. f.). Tree Genet Genomes 4:237–245
Chaix G, Monteuuis O, Garcia C, Alloysius D, Gidiman J, Bacilieri R, Goh DKS (2011) Genetic variation in major phenotypic traits among diverse genetic origins of teak (Tectona grandis L.f.) planted in Taliwas, Sabah, East Malaysia. Ann For Sci 68:1015–1026
Changtragoon S (2001) Forest genetic resources of Thailand: status and conservation. In: Uma Shaanker R, Ganeshaiah KN, Bawa KS (eds) Forest genetic resources: status, threats, and conservation strategies. Oxford and IBH Publishing, New Delhi, pp 141–151
Changtragoon S, Szmidt AE (2000) Genetic diversity of teak (Tectona grandis Linn. f.) in Thailand revealed by random amplified polymorphic DNA (RAPD). In: IUFRO working party 2.08.01 (eds) Tropical species breeding and genetic resources. Forest Genetics for the Next Millennium, Durban, pp 82-83
Danish/FAO Forest Tree Seed Centre (1971) Circular letter no. 1. Provenance testing of teak. Danida Forest Seed Centre. DK3050 Humlebæk, Denmark
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ersts PJ [Internet] Geographic Distance Matrix Generator(version 1.2.3). American Museum of Natural History, Center for Biodiversity and Conservation. Available from http://biodiversityinformatics.amnh.org/open_source/gdmg. (Accessed June 20 2014)
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial-DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Fofana IJ, Lidah YJ, Diarrassouba N, N’guetta SPA, Sangare A, Verhaegen D (2008) Genetic structure and conservation of Teak (Tectona grandis) plantations in Cote d’Ivoire, revealed by site specific recombinase (SSR). Trop Conserv Sci 1:279–292
Fofana IJ, Ofori D, Poitel M, Verhaegen D (2009) Diversity and genetic structure of teak (Tectona grandis L.f) in its natural range using DNA microsatellite markers. New Forest 37:175–195
Graudal L, Kjær ED, Thomsen A, Larsen AB (1997) Planning national programmes for conservation of forest genetic resources. Danida Forest Seed Centre, Technical Note No 48. Danida Forest Seed Centre. http://curis.ku.dk/ws/files/20710949/tn48.pdf. (Accessed February 11 2014)
Graudal L, Kjær ED, Suangtho V, Kaosa-ard A, Sa-ardarvut P (1999) Conservation of forest genetic resources of teak (Tectona grandis L.f.) in Thailand. Technical Note No 52. Danida Forest Seed Centre, Humlebæk and Royal Forest Department, Bangkok. http://curis.ku.dk/ws/files/20711135/technical_note_52.pdf. (Accessed February 11 2014)
Guillot G, Estoup A, Mortier F, Cosson JF (2005a) A spatial statistical model for landscape genetics. Genetics 170:1261–1280
Guillot G, Mortier F, Estoup A (2005b) Geneland: a computer package for landscape genetics. Mol Ecol Notes 5:708–711
Gyi KK, Tint K (1998) Management status of natural teak forests. In: White K, Kashio M (eds) Teak for the future. Proceedings of the Second Regional Seminar on Teak, Yangon, Myanmar, 29 May - 3 June 1995, pp. 27-48. Bangkok, Thailand, FAO Regional Office for Asia and the Pacific
Hartl DL, Clark AG (1997) Principles of population genetics, 3rd edn. Sinauer Associates, Inc., Sunderland
Hedegart T (1973) Pollination of teak (Tectona grandis Linn. f.) 2. Silvae Genet 22:124–128
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Holgate P (1966) A mathematical study of the founder principle of evolutionary genetics. J App Prob 3:115–128
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jost L (2008) G(ST) and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Kalinowski ST (2004) Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv Genet 5:539–543
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189
Kalinowski ST (2009) How well do evolutionary trees describe genetic relationships among populations? Heredity 102:506–513
Kaosa-ard A (1981) Teak (Tectona grandis L.f.)—its natural distribution and related factors. Nat Hist Bull Siam Soc 29:55–74
Kaosa-ard A, Suangtho V, Kjær ED (1998) Experience from tree improvement of teak (Tectona grandis) in Thailand. Technical Note No 50, ISSN 0902-3224. Danida Forest Seed Centre, Humlebæk, Denmark
Keiding H, Wellendorf H, Lauridsen EB (1986) Evaluation of an international series of teak provenance trials. Danida Forest Seed Centre, Humlebæk
Keogh RM (1981) Teak provenances of the Caribbean, central America, Venezuela and Colombia. In: Whitmore JL (ed): Proceedings from IUFRO Symposium on wood production in the neotropics via plantations Sep. 8-12, 1980. Held at Institute for Tropical Forestry, Rio Piedras, Puerto Rico. Published by IUFRO, Vienna
Kertadikara AWS, Prat D (1995a) Genetic-structure and mating system in teak (Tectona-Grandis L-F) provenances. Silvae Genet 44:104–110
Kertadikara AWS, Prat D (1995b) Isozyme variation among teak (Tectona-Grandis Lf) provenances. Theor App Genet 90:803–810
Kimura M, Crow JF (1964) Number of alleles that can be maintained in finite population. Genetics 49:725–738
Kjær ED (2005) Genetic aspects of quality teakwood plantations. In: Bhat KM, Nair KKN, Bhat KV, Muralidharan EM, Sharma JK (eds) Quality timber products of teak from sustainable forest management. Kerala Forest Research Institute. India and International Tropical Timber Organization, Yokohama, pp 311–320
Kjær ED, Foster GS (1996) Economic gains from tree improvement of teak. Technical Note No 43, ISSN 0902-3224. The World Bank/Danida Forest Seed Centre, DK-3050 Humlebæk Denmark
Kjær ED, Siegismund HR (1996) Allozyme diversity in two Tanzanian and two Nicaraguan landraces of teak (Tectona grandis L.). For Genet 3:45–52
Kjær ED, Suangtho V (1995) Outcrossing rate of teak (Tectona grandis (L)). Silvae Genet 44:175–177
Kjær ED, Lauridsen EB, Wellendorf H (1995) Second evaluation of an international series of teak provenance trials. Published by Danida Forest Seed Centre, Humlebaek, Denmark, 117p. http://curis.ku.dk/ws/files/20713023/teakii.pdf. (Accessed February 11 2014)
Kjær ED, Siegismund HR, Suangtho V (1996) A multivariate study on genetic variation in teak (Tectona grandis (L)). Silvae Genet 45:361–368
Kjær ED, Kajornsrichon S, Lauridsen EB (1999) Heartwood, calcium and silica content in five provenances of teak (Tectona grandis L.). Silvae Genet 48:1–3
Kjær ED, Kaosa-ard A, Suangtho V (2000) Domestication of teak through tree improvement. Options, possible gains and critical factors. In: Enters T, Nair CT (eds) Site, technology and productivity of teak plantations. FORSPA Publication No. 24/2000, TEAKNET Pub No. 3. pp 161-189
Kollert W, Cherubini L (2012) Teak resources and market assessment 2010. FAO Planted Forests and Trees Working Paper FP/47/E, Rome. http://www.fao.org/forestry/plantedforests/67508@170537/en/. (Accessed October 2 2013)
Koskela J, Vinceti B, Dvorak W, Bush D, Dawson I, Loo J, Kjær ED, Navarro C, Padolina C, Bordács S, Jamnadass R, Graudal L, Ramamonjisoa L (2010) The use and exchange of forest genetic resources for food and agriculture. FAO Commission on Genetic Resources for Food and Agriculture Background study paper No. 44:1–67, FAO, Rome
Lines R (1965) Standardisation of methods for provenance research and testing. Report of N.G. meeting at Pont-á-Mousson 1965. In: XIV IUFRO-congress, section 22, München 1967, pp. 672-718
Lwin O, Hyun JO, Yahya AF (2010) Assessment of teak (Tectona grandis Linn. f.) provenance tests in the Bago Yoma Region, Myanmar. J Korean Forest Soc 99:686–692
Mandal AK, Chawhan PH (2005) Investigations on inheritance of growth and wood properties and their relationships in teak. In: Bhat KM, Nair KKN, Bhat KV, Muralidharan EM, Sharma JK (eds) Quality timber products of teak from sustainable forest management. Kerala Forest Research Institute. India and International Tropical Timber Organization, Yokohama, pp 506–510
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Meirmans PG, Hedrick PW (2011) Assessing population structure: F(ST) and related measures. Mol Ecol Resour 11:5–18
Minn Y (2012) Investigations of genetic variation of teak (Tectona grandis Linn. f.) in Myanmar for conservation and sustainable utilization of genetic resources. PhD thesis, Georg-August-Universität Göttingen
Monteuuis O, Goh DKS, Garcia C, Alloysius D, Gidiman J, Bacilieri R, Chaix G (2011) Genetic variation of growth and tree quality traits among 42 diverse genetic origins of Tectona grandis planted under humid tropical conditions in Sabah, East Malaysia. Tree Genet Genomes 7:1263–1275
Narayanan C, Chawhaan PH, Mandal AK (2009) Inheritance pattern of growth and wood traits in teak (Tectona grandis L.f.). Silvae Genet 58:97–101
Nicodemus A, Nagarajan B, Narayanan C, Varghese M, Subramanian K (2005) RAPD variation in Indian teak populations and its implications for breeding and conservation. In: Bhat KM, Nair KKN, Bhat KV, Muralidharan EM, Sharma JK (eds) Quality timber products of teak from sustainable forest management. Kerala Forest Research Institute. India and International Tropical Timber Organization, Yokohama, pp 321–330
Parthiban KT, Surendran C, Paramathma M, Sasikumar K (2005) Molecular characterization of teak seed sources using RAPD’s. In: Bhat KM, Nair KKN, Bhat KV, Muralidharan EM, Sharma JK (eds) Quality timber products of teak from sustainable forest management. Kerala Forest Research Institute. India and International Tropical Timber Organization, Yokohama, pp 331–337
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in excel. population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pedersen AP, Hansen JK, Mtika JM, Msangi TH (2007) Growth, stem quality and age-age correlations in a teak provenance trial in Tanzania. Silvae Genet 56:142–148
Persson A (1971) Observations from a provenance trial of Tectona grandis Linn. f. at Longuza, Tanga region. Tanzania Silviculture Res Note 22:1–13
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Saitou N, Nei M (1987) The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shrestha MK, Volkaert H, Van der Straeten D (2005) Assessment of genetic diversity in Tectona grandis using amplified fragment length polymorphism markers. Can J Forest Res 35:1017–1022
Soerianegara I, Lemmens RHMJ (1994) Timber trees: major commercial timbers. PROSEA, Bogor
Sreekanth PM, Balasundaran M, Nazeem PA, Suma TB (2012) Genetic diversity of nine natural Tectona grandis L.f. populations of the Western Ghats in Southern India. Conserv Genet 13:1409–1419
Tangmitcharoen S, Owens JN (1997) Floral biology, pollination, pistil receptivity, and pollen tube growth of teak (Tectona grandis Linn f). Ann Bot 79:227–241
The International Plant Names Index (2014). Published on the Internet http://www.ipni.org (Accessed January 31 2014)
Tsoumis G (1991) Science and technology of wood: structure, properties, utilization. Van Nostrand Reinhold, New York
Verhaegen D, Ofori D, Fofana I, Poitel M, Vaillant A (2005) Development and characterization of microsatellite markers in Tectona grandis (Linn. f). Mol Ecol Notes 5:945–947
Verhaegen D, Fofana IJ, Logossa ZA, Ofori D (2010) What is the genetic origin of teak (Tectona grandis L.) introduced in Africa and in Indonesia? Tree Genet Genomes 6:717–733
Wood PJ (1967) Teak planting in Tanzania. Proceedings from FAO world symposium on man-made forests. Document 3:1631–1644. Canberra, Australia
Acknowledgments
The present work is based on the outcome of the joint, international effort made by researchers from India, Thailand, Laos and Denmark in the 1970s, coordinated by Henrik Keiding who passed away in 2013. Without their effort, the present publication would not have been possible. We thank Mr. Wiroj and Mr. Saroj from Royal Forest Department, who are responsible for field trials in Pha Nak Khao and Ngao, Lampang, respectively, and all their staff, for excellent help during the collection of leaf samples. Our thanks also go to Nutthakorn Semsantud and Verapong Suangtho for invaluable help with logistics and collection at this stage. We thank Dr. Mhando and Dr. Richard from Tanzania Forest Research Institute (TAFORI) for kindly providing a provenance sample from the trial in Tanzania. Lars Schmidt assisted in the field sampling and Morten Alban Knudsen compiled the map of the provenances. This research was supported by the Danish International Development Agency (Danida) and facilitated by cooperation through TEAKNET, the International Teak Information Network. Financial support from the German Federal Environmental Foundation (Deutsche Bundesstiftung Umwelt; DBU) is gratefully acknowledged; it allowed the inclusion of samples from Myanmar in this study.
Data archiving statement
The data—consisting of genotypic data for 6 SSRs in 721 individuals from 29 provenances—will be archived in Dryad (http://datadryad.org/).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. G. Vendramin
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Hansen, O.K., Changtragoon, S., Ponoy, B. et al. Genetic resources of teak (Tectona grandis Linn. f.)—strong genetic structure among natural populations. Tree Genetics & Genomes 11, 802 (2015). https://doi.org/10.1007/s11295-014-0802-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-014-0802-5