Abstract
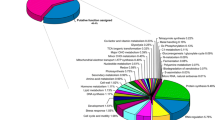
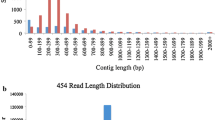
The development of single nucleotide polymorphism (SNP) markers in Japanese pear (Pyrus pyrifolia Nakai) offers the opportunity to use DNA markers for marker-assisted selection in breeding programs because of their high abundance, codominant inheritance, and potential for automated high-throughput analysis. We developed a 1,536-SNP bead array without a reference genome sequence from more than 44,000 base changes on the basis of a large-scale expressed sequence tag (EST) analysis combined with 454 genome sequencing data of Japanese pear ‘Housui’. Among the 1,536 SNPs on the array, 756 SNPs were genotyped, and 609 SNP loci were mapped to linkage groups on a genetic linkage map of ‘Housui’, based on progeny of an interspecific cross between European pear (Pyrus communis L.) ‘Bartlett’ and ‘Housui’. The newly constructed genetic linkage map consists of 951 loci, comprising 609 new SNPs, 110 pear genomic simple sequence repeats (SSRs), 25 pear EST–SSRs, 127 apple SSRs, 61 pear SNPs identified by the “potential intron polymorphism” method, and 19 other loci. The map covers 22 linkage groups spanning 1341.9 cM with an average distance of 1.41 cM between markers and is anchored to reference genetic linkage maps of European pears and apples. A total of 514 contigs containing mapped SNP loci showed significant similarity to known proteins by functional annotation analysis.




Similar content being viewed by others
References
Bell RL, Quamme HA, Layne REC, Skirvin RM (1996) Pears. In: Janick J, Moore JN (eds) Fruit breeding, volume I: tree and tropical fruits. Wiley, New York, pp 441–514
Brookes AJ (1999) The essence of SNPs. Gene 234:177–186
Celton JM, Tustin DS, Chagné D, Gardiner SE (2009a) Construction of a dense genetic linkage map for apple rootstocks using SSRs developed from Malus ESTs and Pyrus genomic sequences. Tree Genet Genomes 5:93–107
Celton JM, Chagné D, Tustin SD, Terakami S, Nishitani C, Yamamoto T, Gardiner SE (2009b) Update on comparative genome mapping between Malus and Pyrus. BMC Research Notes 2:182
Chagné D, Crowhurst RN, Troggio M, Davey MW, Gilmore B, Lawley C, Vanderzande S, Hellens RP, Kumar S, Cestaro A et al (2012) Genome-wide SNP detection, validation, and development of an 8 K SNP array for apple. PLoS One 7:e31745
Chen X, Sullivan PF (2003) Single nucleotide polymorphism genotyping: biochemistry, protocol, cost and throughput. Pharmacogenomics J 3:77–96
Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Muller WEG, Wetter T, Suhai S (2004) Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res 14:1147–1159
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Feltus FA, Wan J, Schulze SR, Estill JC, Jiang N, Paterson AH (2004) An SNP resource for rice genetics and breeding based on subspecies Indica and Japonica genome alignments. Genome Res 14:1812–1819
Fernandez-Fernandez F, Harvey NG, James CM (2006) Isolation and characterization of polymorphic microsatellite markers from European pear (Pyrus communis L.). Mol Ecol Notes 6:1039–1041
Grattapaglia D, Resende MDV (2011) Genomic selection in forest tree breeding. Tree Genet Genomes 7:241–255
Grattapaglia D, Sederoff R (1994) Genetic-linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137:1121–1137
Grattapaglia D, Silva OB, Kirst M, de Lima BM, Faria DA, Pappas GJ (2011) High-throughput SNP genotyping in the highly heterozygous genome of Eucalyptus: assay success, polymorphism and transferability across species. BMC Plant Biol 11:65
Guilford P, Prakash S, Zhu JM, Rikkerink E, Gardiner S, Bassett H, Forster R (1997) Microsatellites in Malus × domestica (apple): abundance, polymorphism and cultivar identification. Theor Appl Genet 94:249–254
Gupta PK, Roy JK, Prasad M (2001) Single nucleotide polymorphisms: a new paradigm for molecular marker technology and DNA polymorphism detection with emphasis on their use in plants. Curr Sci India 80:524–535
Huang XQ, Yang SP, Chinwalla AT, Hillier LW, Minx P, Mardis ER, Wilson RK (2006) Application of a superword array in genome assembly. Nucleic Acids Res 34:201–205
Hummer KE, Janick J (2009) Rosaceae: taxonomy, economic importance, genomics. In: Folta KM, Gardiner SE (eds) Genetics and genomics of Rosaceae. Springer, New York, pp 1–17
Hyten DL, Song Q, Choi IY, Yoon MS, Specht JE, Matukumalli LK, Nelson RL, Shoemaker RC, Young ND, Cregan PB (2008) High-throughput genotyping with the GoldenGate assay in the complex genome of soybean. Theor Appl Genet 116:945–952
Inoue E, Matsuki Y, Anzai H, Evans K (2007) Isolation and characterization of microsatellite markers in Japanese pear (Pyrus pyrifolia Nakai). Mol Ecol Notes 7:445–447
Iwata H, Hayashi T, Terakami S, Takada N, Sawamura Y, Yamamoto T (2013) Potential assessment of genome-wide association study and genomic selection in Japanese pear Pyrus pyrifolia. Breed Sci 63:125–140
Jones E, Chu WC, Ayele M, Ho J, Bruggeman E, Yourstone K, Rafalski A, Smith O, McMullen M, Bezawada C et al (2009) Development of single nucleotide polymorphism (SNP) markers for use in commercial maize (Zea mays L.) germplasm. Mol Breed 24:165–176
Khan MA, Han Y, Zhao YF, Troggio M, Schuyler S, Korban SS (2012) A multi-population consensus genetic map reveals inconsistent marker order among maps likely attributed to structural variations in the apple genome. PLoS One 7:e47864
Kikuchi A (1929) Investigation in 1927 and 1928. 1. Paterclinical incompatibility in Japanese pear. J Okitsu Hort Soc 24:1–6 (in Japanese)
Kim H, Terakami S, Nishitani C, Kurita K, Kanamori H, Katayose Y, Sawamura Y, Saito T, Yamamoto T (2012) Development of cultivar-specific DNA markers based on retrotransposon-based insertional polymorphism in Japanese pear. Breed Sci 62:53–62
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugenic 12:172–175
Kumar S, Chagné D, Bink MCAM, Volz RK, Whitworth C, Carlisle C (2012) Genomic selection for fruit quality traits in apple (Malus × domestica Borkh.). PLoS One 7:e36674
Kumar S, Garrick DJ, Bink MCAM, Whitworth C, Chagné D, Volz RK (2013) Novel genomic approaches unravel genetic architecture of complex traits in apple. BMC Genomics 14:393
Liebhard R, Gianfranceschi L, Koller B, Ryder CD, Tarchini R, Van de Weg E, Gessler C (2002) Development and characterisation of 140 new microsatellites in apple (Malus × domestica Borkh.). Mol Breed 10:217–241
Liebhard R, Koller B, Gianfranceschi L, Gessler C (2003) Creating a saturated reference map for the apple (Malus × domestica Borkh.) genome. Theor Appl Genet 106:1497–1508
Luby JJ, Shaw DV (2001) Does marker-assisted selection make dollars and sense in a fruit breeding program? HortSci 36:872–879
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen ZT et al (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380
Marth GT, Korf I, Yandell MD, Yeh RT, Gu ZJ, Zakeri H, Stitziel NO, Hillier L, Kwok PY, Gish WR (1999) A general approach to single-nucleotide polymorphism discovery. Nat Genet 23:452–456
Martínez-García PJ, Parfitt DE, Ogundiwin EA, Fass J, Chan HM, Ahmad R, Lurie S, Dandekar A, Gradziel TM, Crisosto CH (2013) High density SNP mapping and QTL analysis for fruit quality characteristics in peach (Prunus persica L.). Tree Genet Genomes 9:19–36
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Montanari S, Saeed M, Knäbel M, Kim YK, Troggio M, Malnoy M, Velasco R, Fontana P, Won KH, Durel CE, Perchepied L, Schaffer R, Wiedow C, Bus V, Brewer L, Gardiner SE, Crowhurst RN, Chagné D (2013) Identification of Pyrus single nucleotide polymorphisms (SNPs) and evaluation for genetic mapping in European pear and interspecific Pyrus hybrids. PLoS One 7:e36674
Morin PA, Luikart G, Wayne RK, Grp SW (2004) SNPs in ecology, evolution and conservation. Trends Ecol Evol 19:208–216
Moriya S, Iwanami H, Kotoda N, Haji T, Okada K, Terakami S, Mimida N, Yamamoto T, Abe K (2012) Aligned genetic linkage maps of apple rootstock cultivar ‘JM7’ and Malus sieboldii ‘Sanashi 63’ constructed with novel EST–SSRs. Tree Genet Genomes 8:709–723
Nishitani C, Shimizu T, Fujii H, Terakami S, Yamamoto T (2009a) Analysis of expressed sequence tags from Japanese pear ‘Housui’. Acta Hort 814:645–649
Nishitani C, Terakami S, Sawamura Y, Takada N, Yamamoto T (2009b) Development of novel EST–SSR markers derived from Japanese pear (Pyrus pyrifolia). Breed Sci 59:391–400
Peace C, Bassil N, Main D, Ficklin S, Rosyara UR, Stegmeir T, Sebolt A, Gilmore B, Lawley C, Mockler TC, Bryant DW, Wilhelm L, Iezzoni A (2012) Development and evaluation of a genome-wide 6 K SNP array for diploid sweet cherry and tetraploid sour cherry. PLoS One 7:e48305
Schmid KJ, Sorensen TR, Stracke R, Torjek O, Altmann T, Mitchell-Olds T, Weisshaar B (2003) Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res 13:1250–1257
Shirasawa K, Isobe S, Hirakawa H, Asamizu E, Fukuoka H, Just D, Rothan C, Sasamoto S, Fujishiro T, Kishida Y, Kohara M, Tsuruoka H, Wada T, Nakamura Y, Sato S, Tabata S (2010) SNP discovery and linkage map construction in cultivated tomato. DNA Res 17:381–391
Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP et al (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43:109–116
Silfverberg-Dilworth E, Matasci CL, Van de Weg WE, Van Kaauwen MPW, Walser M, Kodde LP, Soglio V, Gianfranceschi L, Durel CE, Costa F et al (2006) Microsatellite markers spanning the apple (Malus × domestica Borkh.) genome. Tree Genet Genomes 2:202–224
Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc Natl Acad Sci U S A 98:9161–9166
Terakami S, Kimura T, Nishitani C, Sawamura Y, Saito T, Hirabayashi T, Yamamoto T (2009) Genetic linkage map of the Japanese pear ‘Housui’ identifying three homozygous genomic regions. J Jpn Soc Hort Sci 78:417–424
Terakami S, Matsumura Y, Kurita K, Kanamori H, Katayose Y, Yamamoto T, Katayama H (2012) Complete sequence of the chloroplast genome from pear (Pyrus pyrifolia): genome structure and comparative analysis. Tree Genet Genomes 8:841–854
Terakami S, Nishitani C, Yamamoto T (2013) Development of SNP markers for marker-assisted selection in pear. Acta Hort 976:463–469
Van Ooijen JW (2006) JoinMap 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D et al (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–839
Verde I, Bassil N, Scalabrin S, Gilmore B, Lawley CT, Gasic K, Micheletti D, Rosyara UR, Cattonaro F, Vendramin E et al (2012) Development and evaluation of a 9 K SNP array for peach by internationally coordinated SNP detection and validation in breeding germplasm. PLoS One 7:e35668
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223:7–12
Wu WR, Yang L, Jin GL, Zhao XQ, Zheng Y, Xu ZH (2007) PIP: a database of potential intron polymorphism markers. Bioinformatics 23:2174–2177
Wu J, Wang ZW, Shi ZB, Zhang S, Ming R, Zhu SL, Khan MA, Tao ST, Korban SS, Wang H et al (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23:396–408
Yamamoto T, Kimura T, Sawamura Y, Kotobuki K, Ban Y, Hayashi T, Matsuta N (2001) SSRs isolated from apple can identify polymorphism and genetic diversity in pear. Theor Appl Genet 102:865–870
Yamamoto T, Kimura T, Shoda M, Imai T, Saito T, Sawamura Y, Kotobuki K, Hayashi T, Matsuta N (2002) Genetic linkage maps constructed by using an interspecific cross between Japanese and European pears. Theor Appl Genet 106:9–18
Yamamoto T, Kimura T, Hayashi T, Ban Y (2006) DNA profiling of fresh and processed fruits in pear. Breed Sci 56:165–171
Yamamoto T, Kimura T, Terakami S, Nishitani C, Sawamura Y, Saito T, Kotobuki K, Hayashi T (2007) Integrated reference genetic linkage maps of pear based on SSR and AFLP markers. Breed Sci 57:321–329
Yamamoto T, Terakami S, Moriya S, Hosaka F, Kurita K, Kanamori H, Katayose Y, Saito T, Nishitani C (2013) DNA markers developed from genome sequencing analysis in Japanese pear (Pyrus pyrifolia). Acta Hort 976:477–483
Yu HH, Xie WB, Wang J, Xing YZ, Xu CG, Li XH, Xiao JH, Zhang QF (2011) Gains in QTL detection using an ultra-high density SNP map based on population sequencing relative to traditional RFLP/SSR markers. PLoS One 6:e17595
Zhu YL, Song QJ, Hyten DL, Van Tassell CP, Matukumalli LK, Grimm DR, Hyatt SM, Fickus EW, Young ND, Cregan PB (2003) Single-nucleotide polymorphisms in soybean. Genetics 163:1123–1134
Acknowledgments
We thank Ms. F. Hosaka, N. Shigeta, and N. Yagihashi for their technical help. This work was partially supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation, SGE-1003).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Dirlewanger
Data submission
All of the pyrosequence data (.sff files) were deposited in DDBJ Sequence Read Archive (DRA001738).
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLSX 216 kb)
Rights and permissions
About this article
Cite this article
Terakami, S., Nishitani, C., Kunihisa, M. et al. Transcriptome-based single nucleotide polymorphism markers for genome mapping in Japanese pear (Pyrus pyrifolia Nakai). Tree Genetics & Genomes 10, 853–863 (2014). https://doi.org/10.1007/s11295-014-0726-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-014-0726-0