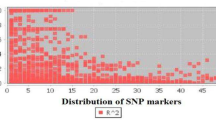
Abstract
We developed a quantitative and association genetic study with Eucalyptus urophylla using a progeny trial. Based on a sample of 831 trees distributed in 84 half-sib families whose wood was phenotyped by near-infrared spectroscopy, the results showed that traits related to lignin, cellulose, and wood extractives presented significant additive genetic variability with moderate to high narrow sense heritability (h 2 = 0.28 to 0.93). Genetic correlations varied with high standard error and showed low to moderate values. Using three cellulose synthase genes (EuCesA1, EuCesA2, and EuCesA3) and three candidate genes involved in the lignin pathway (EuC4H1, EuC4H2, and EuCAD2), an association study was performed for each of the gene action models (co-dominant, recessive, and dominant) using two methods. Firstly, single-marker association tests were done and 539 tests (49 single nucleotide polymorphisms (SNPs) × 11 traits) were analyzed. After Bonferroni correction with a significance level of P = 0.00102, only four SNPs presented significant association with syringyl and syringyl-to-guaiacyl ratio with an adjusted coefficient of determination varying between 2.6 and 4.4 %. Secondly, a model selection method, the backward approach, was implemented. Similar SNPs were detected by both the backward selection and the individual marker approaches. However, the latter detected new associations with other traits, genes, and SNPs and improved the quality of the model as shown by the BIC criteria and the higher adjusted determination coefficient (1.5 to 8.3 %). Our results reveal that cellulose genes can be associated with lignin traits (syringyl-to-guaiacyl ratio) and stress the possible pleiotropic effect of some genes.



Similar content being viewed by others
References
Apiolaza LA, Raymond CA, Yeo BJ (2005) Genetic variation of physical and chemical wood properties of Eucalyptus globulus. Silvae Genet 54:160–166
Ayers KL, Cordell HJ (2010) SNP selection in genome-wide and candidate gene studies via penalized logistic regression. Genet Epidemiol 34:879–891
Barratt BJ, Payne F, Lowe CE, Hermann R, Healy BC, Harold D, Concannon P, Gharani N, McCarthy MI, Olavensen MG, McCormack R, Guja C, Ionescu-Tirgoviste C, Undlien DE, Ronningen KS, Gillespie KM, Tuomilehto-Wolf E, Tuomilehto J, Benett ST, Clayton DG (2004) Remapping the insulin gene/IDDM2 locus in type 1 diabetes. Diabetes 53:1884–1889
Beaulieu J, Doerksen T, Boyle B, Clement S, Deslauriers M, Beauseigle S, Blais S, Poulin PL, Lenz P, Caron S et al (2011) Association genetics of wood physical traits in the conifer white spruce and relationships with gene expression. Genetics 188:197–214
Bernardo R (2008) Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci 48:1649–1664
Bousquet SL, Simon L, Lalonde M (1990) DNA amplification from vegetative and sexual tissue of trees using polymerase chain reaction. Can J For Res 20:254–257
Bouvet J-M, Vigneron P, Villar E, Saya A (2009a) Determining the optimal age for selection by modelling the age-related trends in genetic parameters in Eucalyptus hybrid populations. Silvae Genet 58:102–112
Bouvet J-M, Saya A, Vigneron P (2009b) Trends in additive, dominance and environmental effects with age for growth traits in Eucalyptus hybrid populations. Euphytica 165:35–54
Brondani RPV, Brondani C, Tarchini R, Grattapaglia D (1998) Development, characterization and mapping of microsatellite markers in Eucalyptus grandis and E. urophylla. Theor Appl Genet 97:816–827
Browning BL, Browning SR (2009) A unified approach to genotype imputation and haplotype phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84:210–223
Bush D, McCarthy K, Meder R (2011) Genetic variation of natural durability traits in Eucalyptus cladocalyx (sugar gum). Ann For Sci 68:1057–1066
Calus MPL, Veerkamp RF, Mulder HA (2011) Imputation of missing single nucleotide polymorphism genotypes using a multivariate mixed model framework. J Anim Sci 89:2042–2049
Campbell MM, Sederoff RR (1996) Variation in lignin content and composition—mechanisms of control and implications for the genetic improvement of plants. Plant Physiol 110(3–1):3
Cano-Delgado A, Penfield S, Smith C, Catley M, Bevan M (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34:351–362
Cho S, Kim K, Kim YJ, Lee J-K, Cho YS, Lee J-Y, Han B-G, Kim H, Ott J, Park T (2010) Joint identification of multiple genetic variants via elastic-net variable selection in a genome-wide association analysis. Ann Hum Genet 74:416–428
Cordell HJ, Clayton DG (2002) A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am J Hum Genet 70:124–141
Costa e Silva J, Borralho NMG, Araújo JA, Vaillancourt RE, Potts BM (2009) Genetic parameters for growth, wood density and pulp yield in Eucalyptus globulus. Tree Genet Genomes 5:291–305
Cumbie WP, Eckert A, Wegrzyn J, Whetten R, Neale D, Goldfarb B (2011) Association genetics of carbon isotope discrimination, height and foliar nitrogen in a natural population of Pinus taeda L. Heredity 107:105–114
de Assis TF (2000) Production and use of Eucalyptus hybrids for industrial purposes. In: Nikles DG (ed) Hybrid breeding and genetics of forest trees. Proceedings of QFRI/CRC-SPF Symposium, 9–14th April 2000 Noosa, Queensland, Australia. Department of Primary Industries, Brisbane, pp 63–74
Denis M, Bouvet J-M (2012) Efficiency of genomic selection with models including dominance effect in the context of Eucalyptus breeding. Tree Genet Genomes. doi:10.1007/s11295-012-0528-1
Dillon SK, Brawner JT, Meder R, Lee DJ, Southerton SG (2012) Association genetics in Corymbia citriodora subsp. variegata identifies single nucleotide polymorphisms affecting wood growth and cellulosic pulp yield. New Phytol. doi:10.1111/j.1469-8137.2012.04200.x
Dmitriev DA, Rakitov RA (2008) Onwards Indelligent v.1.2. Http://ctap.inhs.uiuc.edu/dmitriev/indel.asp
Efroymson MA (1960) Multiple regression analysis. In: Ralston A, Wilf HS (eds) Mathematical methods for digital computers. Wiley, New York
Fornalé S, Capellades M, Encina A, Wang K, Irar S, Lapierre C, Ruel K, Joseleau J-P, Berenguer J, Puigdomènech P, Rigau J, Caparrós-Ruiz D (2011) Altered lignin biosynthesis improves cellulosic bioethanol production in transgenic maize plants down-regulated for cinnamyl alcohol dehydrogenase. Mol Plant 5:817–830
Freeman JS, Whittock SP, Potts BM, Vaillancourt RE (2009) QTL influencing growth and wood properties in Eucalyptus globules. Tree Genet Genomes 5:713–722
Fries A, Ericsson T, Gref R (2000) High heritability of wood extractives in Pinus sylvestris progeny tests. Can J For Res 30:1707–1713
Gion J-M, Carouché A, Deweer S, Bedon F, Pichavant F, Charpentier J-P, Baillères H, Rozenberg P, Carocha V, Ognouabi N, Verhaegen D, Grima-Pettenati J, Vigneron P, Plomion C (2011) Comprehensive genetic dissection of wood properties in a widely-grown tropical tree: Eucalyptus. BMC Genom 12:301
Grattapaglia D, Kirst M (2008) Eucalyptus applied genomics: from gene sequences to breeding tools. New Phytol 179:911–929
Grattapaglia D, Vaillancourt R, Merv S, Thumma BR, Foley W, Külheim C, Potts BM, Myburg AA (2012) Progress in Myrtaceae genetics and genomics: Eucalyptus as the pivotal genus. Tree Genet Genomes. doi:10.1007/s11295-012-0491-x
Gonzalez-Martinez SC, Wheeler NC, Ersoz E, Nelson CD, Neale DB (2007) Association genetics in Pinus taeda L. I. Wood property traits. Genetics 175:399–409
Gilmour AR, Gogel BJ, Cullis BR, Welham SJ, Thompson R (2005) ASReml user guide release 2.0. VSN International Ltd, Hemel Hempstead HP1 1ES
Hamilton MG, Raymond CA, Harwood CE, Potts BM (2009) Genetic variation in Eucalyptus nitens pulpwood and wood shrinkage traits. Tree Genet Genomes 5:307–316
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population level. Mol Ecol Notes 2:618–620
Hein PRG, Lima JT, Chaix G (2009) Robustness of models based on near infrared spectra to predict the basic density in Eucalyptus urophylla wood. J Near Infrared Spec 17:141–150
Hein PRG, Lima JT, Chaix G (2010) Effects of sample preparation on NIR spectroscopic estimation of chemical properties of Eucalyptus urophylla ST Blake wood. Holzforschung 64:45–54
Hein PRG, Bouvet J-M, Mandrou E, Vigneron P, Clair B, Chaix G (2012) Age trends of microfibril angle inheritance and their genetic and environmental correlations with growth, density and chemical properties in Eucalyptus urophylla ST Blake wood. Ann For Sci. doi:10.1007/s13595-012-0186-3
Hoggart CJ, John C, Whittaker JC, De Iorio M, Balding DJ (2008) Simultaneous analysis of all SNPs in genome-wide and re-sequencing association studies. PLoS Genetics 4:e1000130
Hull J, Campino S, Rowlands K, Chan M-S, Copley RR, Taylor MS, Rockett K, Elvidge G, Keating B, Knight J, Kwiatkowski D (2007) Identification of common genetic variation that modulates alternative splicing. PLoS Genet 3(6):e99
Ingvarsson PK, Garcia MV, Luquez V, Hall D, Jansson S (2008) Nucleotide polymorphism and phenotypic associations within and around the phytochrome b2 locus in European aspen (Populus tremula, Salicaceae). Genetics 178:2217–2226
Keegstra K (2010) Plant cell walls. Plant Physiol 154:483–486
Kien ND, Quang TH, Jansson G, Harwood C, Clapham D, von Arnold S (2009) Cellulose content as a selection trait in breeding for kraft pulp yield in Eucalyptus urophylla. Ann For Sci 66:711
Külheim C, Yeoh SH, Wallis IR, Laffan S, Moran GF, Foley WJ (2011) The molecular basis of quantitative variation in foliar secondary metabolites in Eucalyptus globulus. New Phytol 191:1041–1053
Lauvergeat V, Rech P, Jauneau A, Guez C, Coutos-Thevenot P, Grima-Pettenati J (2002) The vascular expression pattern directed by the Eucalyptus gunnii cinnamyl alcohol dehydrogenase EgCAD2 promoter is conserved among woody and herbaceous plant species. Plant Mol Biol 50:497–509
Ma X-F, Hall D, St Onge KR, Stefan J, Ingvarsson PK (2010) Genetic differentiation, clinal variation and phenotypic associations with growth cessation across the Populus tremula photoperiodic pathway. Genetics 186:1033–1044
Mandrou E, Hein PRG, Villar E, Vigneron P, Plomion C, Gion J-M (2012) A candidate gene for lignin composition in Eucalyptus: cinnamoyl–CoA reductase (CCR). Tree Genet Genomes. doi:10.1007/s11295-011-0446-7
Martin B, Cossalter C (1976a) Les Eucalyptus des îles de la Sonde Partie 1 Bois et Forêt des Tropiques 165: 3-20
Martin B, Cossalter C (1976b) Les Eucalyptus des îles de la Sonde Partie 2 Bois et Forêt des Tropiques 166: 3-22
Martin B, Cossalter C (1976c) Les Eucalyptus des îles de la Sonde Partie 3 Bois et Forêt des Tropiques 167: 3-24
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Mimms A, Kuckurek MJ, Pyiatte JA, Wright EE (1993) Kraft pulping. A compilation of notes. TAPPI, Atlanta, pp 6–7
Mrode RA, Thompson R (2005) Linear models for the prediction of animal breeding values, 2nd edn. CABI, Cambridge
Neale DB, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12:111–122
Payn KG, Dvorak WS, Janse BJH, Myburg AA (2008) Microsatellite diversity and genetic structure of the commercially important tropical tree species Eucalyptus urophylla, endemic to seven islands in eastern Indonesia. Tree Genet Genome 4:519–530
Poke FS, Potts BM, Vaillancourt RE, Raymond CA (2006) Genetic parameters for lignin, extractives and decay in Eucalyptus globulus. Ann For Sci 63:813–821
Pritchard JK, Falush D, Wen X (2007) Documentation for structure software, version 22. http://pritch.bsdu.chicago.edu.
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Quang TH, Duc KN, von Arnold S, Jansson G, Thinh HH, Clapham D (2010) Relationship of wood composition to growth traits of selected open-pollinated families of Eucalyptus urophylla from a progeny trial in Vietnam. New For 39:301–312
R Development Core Team (2011) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Raymond CA (2002) Genetics of Eucalyptus wood properties. Ann For Sci 59:525–531
Ranik M, Myburg AA (2006) Six new cellulose synthase genes from Eucalyptus are associated with primary and secondary cell wall biosynthesis. Tree Physiol 26:545–556
Rao DC (2008) An overview of the genetic dissection of complex traits. Adv Genet 60:3–34
Resende MFRJ, Munoz P, Acosta JJ, Peter GF, Davis JM, Grattapaglia D, Resende MDV, Kirst M (2012) Accelerating the domestication of trees using genomic selection: accuracy of prediction models across ages and environments. New Phytol 193:617–624
Rogers LA, Campbell MMC (2004) The genetic control of lignin deposition during plant growth and development. New Phytol 164:17–30
Schwarz GE (1978) Estimating the dimension of a model. Ann Stat 6:461–464, 2010
Southerton SG, MacMillan CP, Bell JC, Bhuiyan N, Downes G, Ravenwood IC, Joyce KR, Williams D, Thumma BR (2010) Association of allelic variation in xylem genes with wood properties in Eucalyptus nitens. Aust Forest J 73:259–264
Stackpole DJ, Vaillancourt RE, Alves A, Rodrigues J, and Potts BM (2011) Genetic variation in the chemical components of Eucalyptus globulus wood. G3 1: 151-159
Stich B, Möhring J, Piepho H-P, Heckenberger M, Buckler ES, Melchinger AE (2008) Comparison of mixed-model approaches for association mapping. Genetics 178:1745–1754
Storey JD, Tibshirani R (2003) Statistical significance for genome-wide experiments. Proc Natl Acad Sci USA 100:9440–9445
Storey JD, Taylor JE, Siegmund D (2004) Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: a unified approach. J R Stat Soc B 66:187–205
Sykes R, Li B, Isik F, Kadla J, Chang H-M (2006) Genetic variation and genotype by environment interactions of juvenile wood chemical properties in Pinus taeda L. Ann For Sci 63:897–904
Sun G, Zhu C, Kramer MH, Yang S-S, Song W, Piepho H-P, Yu J (2010) Variation explained in mixed-model association mapping. Heredity 105:333–340
Tibbits W, Hodge G (1998) Genetic parameters and breeding value predictions for Eucalyptus nitens wood fiber production traits. For Sci 44:587–598
Thumma BR, Nolan MR, Evans R, Moran GF (2005) Polymorphisms in cinnamoyl CoA reductase (CCR) are associated with variation in microfibril angle in Eucalyptus spp. Genetics 171:1257–1265
Thumma BR, Matheson BA, Zhang DQ, Meeske C, Meder R, Downes GM, Southerton SG (2009) Identification of a cis-acting regulatory polymorphism in a eucalypt cobra-like gene affecting cellulose content. Genetics 183:1153–1164
Thumma BR, Southerton SG, Bell JC, Owen JV, Henery ML, Moran GF (2010) Quantitative trait locus (QTL) analysis of wood quality traits in Eucalyptus nitens. Tree Genet Genomes 6:305–317
Tripiana V, Bourgeois M, Verhaegen D, Vigneron P, Bouvet JM (2007) Combining microsatellites, growth, and adaptive traits for managing in situ genetic resources of Eucalyptus urophylla. Can J For Res 37:773–785
Verryn SD (2000) Eucalyptus hybrid breeding in South Africa. In: Nikles DG (ed) Hybrid breeding and genetics of forest trees. Proceedings of QFRI/CRC-SPF Symposium, 9–14th April 2000 Noosa, Queensland, Australia. Department of Primary Industries, Brisbane, pp 191-199
Wang KS, Mullersman JE, Liu XF (2010) Family-based association analysis of the MAPT gene in Parkinson disease. J Appl Genet 51:509–514
Wegrzyn JL, Eckert AJ, Choi M, Lee JM, Stanton BJ, Sykes R, Davis MF, Tsai C-J, Neale DB (2010) Association genetics of traits controlling lignin and cellulose biosynthesis in black cottonwood (Populus trichocarpa, Salicaceae) secondary xylem. New Phytol 188:515–532
Werck-Reichhart D, Feyereisen R (2000) Cytochromes P450: a success story. Genome Biol 1:3003.1–3003.9
Xu Z, Zhang D, Hu J, Zhou X, Ye X, Reichel KL, Stewart NR, Syrenne RD, Yang X, Gao P, Shi W, Doeppke C, Sykes RW, Burris JN, Bozell JJ, Cheng Z-M, Hayes DG, Labbe N, Davis M, Stewart CN, Yuan JS (2009) Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinforma 10:S3
Yu Q, Pulkkinen P, Rautio M, Haapanen M, Alén R, Stener LG, Beuker E, Tigerstedt PMA (2001) Genetic control of wood physicochemical properties, growth, and phenology in hybrid aspen clones. Can J For Res 31:1348–1356
Yu JG, Briggs WH, Bi IV, Yamasaki M et al (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Zhang D, Zhang Z, Yang K (2006) QTL analysis of growth and wood chemical content traits in an interspecific backcross family of white poplar (Populus tomentosa × P. bolleana) × P. tomentosa. Can J For Res 36:2015–2023
Zobel BJ, van Buijtenen JP (1989) Wood variation. Its causes and control. Springer, Heidelberg
Acknowledgments
We are grateful to CIRAD and especially to the three founders of the research center “Centre de Recherche sur le Durabilité et la Productivité des Plantations Industrielles” (CRDPI) of the Republic of the Congo in Pointe Noire for having supported part of this research over several years. We also thank the Ministry of Research of the Republic of the Congo, the company EFC, and CIRAD. We are grateful to the monitoring team of CRDPI for providing information and data on the experimental design.
Data Archiving Statement
Phenotypic data
SNP data
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Grattapaglia
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 18 kb)
ESM 2
(DOCX 27 kb)
ESM 3
(DOCX 20 kb)
ESM 4
(DOCX 22 kb)
ESM 5
(DOCX 24 kb)
Fig. S 6
Illustration of the association test for the 11 growth and wood traits and the 49 SNPs and the three gene action models (co-dominant, recessive, and dominant) (PPTX 66 kb)
ESM 7
(PPTX 85 kb)
ESM 8
(PPTX 84 kb)
ESM 9
(PPTX 84 kb)
ESM 10
(PPTX 83 kb)
ESM 11
(PPTX 83 kb)
Rights and permissions
About this article
Cite this article
Denis, M., Favreau, B., Ueno, S. et al. Genetic variation of wood chemical traits and association with underlying genes in Eucalyptus urophylla . Tree Genetics & Genomes 9, 927–942 (2013). https://doi.org/10.1007/s11295-013-0606-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-013-0606-z