Abstract
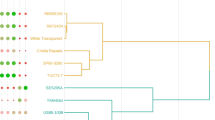
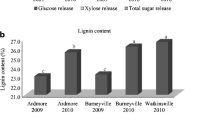
We have measured significant genetically determined variation in biomass composition among breeding populations of shrub willow, a biomass feedstock crop. This project was aimed to ask whether patterns of cell wall gene expression can be correlated with genetic variation in biomass composition at harvest, in order to develop assays of early differences in gene expression as indicators of harvestable biomass chemical composition and potentially reduce the time of selection for new willow genotypes. Previous studies have demonstrated that manipulation of expression of cell wall biosynthetic genes results in altered biomass chemical composition. We analyzed genes encoding enzymes involved in lignin biosynthesis and carbohydrate active enzymes selected based on their functional characterization and conservation in Populus trichocarpa and Arabidopsis thaliana. Fragments of 20 genes were cloned from young stem cDNA of Salix sachalinensis and Salix miyabeana. Expression profiling in willow stem apical tissue and developing stem tissue was performed for each isolated gene using probe-based quantitative real-time PCR. Two willow parental genotypes and six progeny within a hybrid family were selected for analysis, and significant differences in expression among the individuals and between tissue types were observed for most of the genes. Significant correlations between patterns of gene expression and variation in the biomass chemical composition of those genotypes provide insight into the genetic regulation of lignocellulosic deposition in this important bioenergy crop and could be utilized as a tool for early selection of new genotypes.






Similar content being viewed by others
References
Aspeborg H, Schrader J, Coutinho PM, Stam M, Kallas A, Djerbi S, Nilsson P, Denman S, Amini B, Sterky F, Master E, Sandberg G, Mellerowicz E, Sundberg B, Henrissat B, Teeri TT (2005) Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol 137:983–997
Atanassov II, Pittman JK, Turner SR (2009) Elucidating the mechanisms of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. J Biol Chem 284:3833–3841
Barakat A, Bagniewska-Zadworna A, Choi A, Plakkat U, DiLoreto D, Yellanki P, Carlson J (2009) The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression. BMC Plant Biol 9:26
Baucher M, Chabbert B, Pilate G, Van Doorsselaere J, Tollier M, Petit-Conil M, Cornu D, Monties B, Van Montagu M, Inze D, Jouanin L, Boerjan W (1996) Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol 112:1479–1490
Baucher M, Halpin C, Petit-Conil M, Boerjan W (2003) Lignin: genetic engineering and impact on pulping. Crit Rev Biochem Mol Biol 38:305–350
Bergmann S, Ihmels J, Barkai N (2003) Similarities and differences in genome-wide expression data of six organisms. PLoS Biol 2:85–93
Boerjan W (2005) Biotechnology and the domestication of forest trees. Curr Opin Biotechnol 16:159–166
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Ann Rev Plant Biol 54:519–546
Broekgaarden C, Poelman E, Voorrips R, Dicke M, Vosman B (2010) Intraspecific variation in herbivore community composition and transcriptional profiles in field-grown Brassica oleracea cultivars. J Exp Bot 61:807–819
Brown DM, Zhang Z, Stephens E, Dupree P, Turner SR (2009) Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J 57:732–746
Brunner A, Yakovlev I, Strauss S (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4:14
Cameron KD, Kopp RF, Abrahamson LP, Volk TA, Smart LB (2010) High-yielding shrub willow (Salix spp.) emerges as a robust bioenergy crop suitable for growth in the Northeast, Upper Midwest, and Southern Canada. http://abstracts.aspb.org/pb2010/public/P01/P01054.html. American Society of Plant Biologists—Plant Biology 2010, Montreal, Canada
Campbell MM, Sederoff RR (1996) Variation in lignin content and composition (mechanisms of control and implications for the genetic improvement of plants). Plant Physiol 110:3–13
Carroll S (2000) Endless forms: the evolution of gene regulation and morphological diversity. Cell 101:577–580
Carter MJ, Milton ID (1993) An inexpensive and simple method for DNA purifications on silica particles. Nucleic Acids Res 21:1044
Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25:759–761
Coutinho PM, Stam M, Blanc E, Henrissat B (2003) Why are there so many carbohydrate-active enzyme-related genes in plants? Trends Plant Sci 8:563–565
Dauwe R, Morreel K, Goeminne G, Gielen B, Rohde A, Beeumen JV, Ralph J, Boudet A-M, Kopka J, Rochange SF, Halpin C, Messens E, Boerjan W (2007) Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J 52:263–285
Day A, Neutelings G, Nolin F, Grec S, Habrant A, Cronier D, Mahar B, Rolando C, David H, Chabbert B, Hawkins S (2009) Caffeoyl coenzyme A O-methyltransferase down-regulation is associated with modifications in lignin and cell-wall architecture in flax secondary xylem. Plant Physiol Biochem 47:9–19
Dharmawardhana P, Brunner A, Strauss S (2010) Genome-wide transcriptome analysis of the transition from primary to secondary stem development in Populus trichocarpa. BMC Genomics 11:150
Dinus RJ, Payne P, Sewell MM, Chiang VL, Tuskan GA (2001) Genetic modification of short rotation poplar wood: properties for ethanol fuel and fiber productions. Crit Rev Plant Sci 20:51–69
Doebley J, Lukens L (1998) Transcriptional regulators and the evolution of plant form. Plant Cell 10:1075–1082
Elissetche JP, Valenzuela S, Garcia R, Norambuena M, Iturra C, Rodriguez J, Mendonca RT, Balocchi C (2011) Transcript abundance of enzymes involved in lignin biosynthesis of Eucalyptus globulus genotypes with contrasting levels of pulp yield and wood density. Tree Genet Genome 7:697–705
Friedmann M, Ralph SG, Aeschliman D, Zhuang J, Ritland K, Ellis BE, Bohlmann J, Douglas CJ (2007) Microarray gene expression profiling of developmental transitions in Sitka spruce (Picea sitchensis) apical shoots. J Exp Bot 58:593–614
Geisler-Lee J, Geisler M, Coutinho PM, Segerman B, Nishikubo N, Takahashi J, Aspeborg H, Djerbi S, Master E, Andersson-Gunneras S, Sundberg B, Karpinski S, Teeri TT, Kleczkowski LA, Henrissat B, Mellerowicz EJ (2006) Poplar carbohydrate-active enzymes. Gene identification and expression analysis. Plant Physiol 140:946–962
González-Martínez SC, Wheeler NC, Ersoz E, Nelson CD, Neale DB (2007) Association genetics in Pinus taeda L. I. wood property traits. Genetics 175:399–409
Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Himmel ME, Ding S-Y, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD (2007) Biomass recalcitrance: engineering plants and enzymes for biofuel production. Science 315:804–807
Hu W, Harding S, Lung J, Popko J, Ralph J, Stokke D, Tsai C, Chiang V (1999) Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol 17:808–812
Johansson H, Sterky F, Amini B, Lundenberg J, Kleczkowski LA (2002) Molecular cloning and characterization of a cDNA encoding poplar UDP-glucose dehydrogenase, a key gene of hemicellulose/pectin formation. Biochim Biophys Acta Gene Struct Expr 1576:53–58
Joshi CP, Bhandari S, Ranjan P, Kalluri U, Liang X, Fujino T, Samuga A (2004) Genomics of cellulose biosynthesis in poplars. New Phytol 164:53–61
Jouanin L, Goujon T, de Nadai V, Martin M-T, Mila I, Vallet C, Pollet B, Yoshinaga A, Chabbert B, Petit-Conil M, Lapierre C (2000) Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol 123:1363–1374
Kirst M, Basten CJ, Myburg AA, Zeng Z-B, Sederoff RR (2005) Genetic architecture of transcript-level variation in differentiating xylem of a Eucalyptus hybrid. Genetics 169:2295–2303
Klinghammer M, Tenhaken R (2007) Genome-wide analysis of the UDP-glucose dehydrogenase gene family in Arabidopsis, a key enzyme for matrix polysaccharides in cell walls. J Exp Bot 58:3609–3621
Kumar M, Thammannagowda S, Bulone V, Chiang V, Han K-H, Joshi CP, Mansfield SD, Mellerowicz E, Sundberg B, Teeri T, Ellis BE (2009) An update on the nomenclature for the cellulose synthase genes in Populus. Trends Plant Sci 14:248–254
Lapierre C, Pollet B, Petit-Conil M, Toval G, Romero J, Pilate G, Leple J, Boerjan W, Ferret V, De Nadai V, Jouanin L (1999) Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial Kraft pulping. Plant Physiol 119:153–164
Liang X, Joshi CP (2004) Molecular cloning of ten distinct hypervariable regions from the cellulose synthase gene superfamily in aspen trees. Tree Physiol 24:543–550
Machabo H, Cruzan M (2010) Intraspecific variation in gene expression under prolonged drought in Piriqueta hybrids and their parental taxa. Plant Sci 178:429–439
Mészáros E, Jakab E, Várhegyi G, Szepesváry P, Marosvölgyi B (2004) Comparative study of the thermal behavior of wood and bark of young shoots obtained from an energy plantation. J Anal Appl Pyrol 72:317–328
Meyermans H, Morreel K, Lapierre C, Pollet B, De Bruyn A, Busson R, Herdewijn P, Devreese B, Van Beeumen J, Marita JM, Ralph J, Chen C, Burggraeve B, Van Montagu M, Messens E, Boerjan W (2000) Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J Biol Chem 275:36899–36909
Orfao JJM, Antunes FJA, Figueiredo JL (1999) Pyrolysis kinetics of lignocellulosic materials—three independent reactions model. Fuel 78:349–358
Orfila C, Sorensen S, Harbolt J, Geshi N, Crombie H, Truong H-N, Reid J, Knox J, Scheller H (2005) Quasimodo1 is expressed in vascular tissue of Arabidopsis thaliana inflorescence stems, and affects homogalacturonan and xylan biosythesis. Planta 222:613–622
Pilate G, Guiney E, Holt K, Petit-Conil M, Lapierre C, Leple J, Pollet B, Mila I, Webster E, Marstorp H, Hopkins D, Jouanin L, Boerjan W, Schuch W, Cornu D, Halpin C (2002) Field and pulping performances of transgenic trees with altered lignification. Nat Biotechnol 20:607–612
Ralph J, Mackay JJ, Hatfield R, O'Malley D, Whetten R, Sederoff RR (1997) Abnormal lignin in a loblolly pine mutant. Science 277:235–239
Ranik M, Myburg AA (2006) Six new cellulose synthase genes from Eucalyptus are associated with primary and secondary cell wall biosynthesis. Tree Physiol 26:545–556
Ranjan P, Yin T, Zhang X, Kalluri U, Yang X, Jawdy S, Tuskan G (2010) Bioinformatics-based identification of candidate genes from QTLs associated with cell wall traits in Populus. Bioenerg Res 3:172–182
Raveendran K, Ganesh A, Khilar KC (1996) Pyrolysis characteristics of biomass and biomass components. Fuel 75:987–998
Samac DA, Litterer L, Temple G, Jung H, Somers DA (2004) Expression of UDP-glucose dehydrogenase reduces cell-wall polysaccharide concentration and increases xylose content in alfalfa stems. Appl Biochem Biotech 113–116:1167–1182
SAS Institute Inc. SAS 9.1.3 Help and Documentation. Cary, NC: SAS Institute Inc., 2000–2004
Serapiglia MJ, Cameron KD, Stipanovic AJ, Smart LB (2008) High-resolution thermogravimetric analysis for rapid characterization of biomass composition and selection of shrub willow varieties. Appl Biochem Biotechnol 145:3–11
Serapiglia MJ, Cameron KD, Stipanovic AJ, Smart LB (2009) Analysis of biomass composition using high-resolution thermogravimetric analysis and percent bark content as tools for the selection of shrub willow bioenergy crop varieties. Bioenerg Res 2:1–9
Shafizadeh F, Chin PPS (1977) Thermal decomposition of wood. In: Goldstein IS (ed) Wood technology: chemical aspects, pp. 57–81. American Chemical Society Symposium Series 43
Shi R, Sun Y-H, Heber S, Sederoff R, Chiang V (2010) Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol 51:144–163
Singh K, Risse M, Das KC, Worley J (2009) Determination of composition of cellulose and lignin mixtures using thermogravimetric analysis. J Energ Resour Tech 131:022201–022206
Song D, Shen J, Li L (2010) Characterization of cellulose synthase complexes in Populus xylem differentiation. New Phytol 187:777–790
Stern DL (2000) Perspective: evolutionary developmental biology and the problem of variation. Evolution 54:1079–1091
Sticklen M (2006) Plant genetic engineering to improve biomass characteristics for biofuels. Curr Opin Biotechnol 17:315–319
Stipanovic AJ, Goodrich J, Hennessy P (2004) High resolution thermogravimetric analysis (HR-TGA) in the compositional characterization of polysaccharides American Chemical Society symposium on “novel analytical tools in the characterization of polysaccharides”. Cellulose and Renewable Materials Division, Philadelphia
Thumma BR, Southerton SG, Bell JC, Owen JV, Henery ML, Moran GF (2010) Quantitative trait locus (QTL) analysis of wood quality traits in Eucalyptus nitens. Tree Genet Genome 6:305–317
Tsai C-J, Harding SA, Tshaplinski TJ, Lindroth RL, Yuan Y (2006) Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol 172:47–62
Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen G-L, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Dejardin A, dePamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M, Jorgensen R, Joshi C, Kangasjarvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Leple J-C, Locascio P, Lou Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson O, Pereda V, Peter G, Philippe R, Pilate G, Poliakov A, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouze P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai C-J, Uberbacher E, Unneberg P et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Vanholme R, Morreel K, Ralph J, Boerjan W (2008) Lignin engineering. Curr Opin Biotechnol 11:278–285
Voelker SL, Lachenbruch B, Meinzer FC, Jourdes M, Ki C, Patten AM, Davin LB, Lewis NG, Tuskan GA, Gunter L, Decker SR, Selig MJ, Sykes R, Himmel ME, Kitin P, Shevchenko O, Strauss SH (2010) Antisense down-regulation of 4CL expression alters lignification, tree growth, and saccharification potential of field-grown poplar. Plant Physiol 154:874–886
Wagner A, Donaldson L, Kim H, Phillips IS, Flint H, Steward D, Torr K, Koch G, Schmitt U, Ralph J (2009) Suppression of 4-coumarate-CoA ligase in the coniferous gymnosperm Pinus radiata. Plant Physiol 149:370–383
Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223:7–12
Wang L, Weller CL, Jones DD, Hanna MA (2008) Contemporary issues in thermal gasification of biomass and its application to electricity and fuel production. Biomass Bioenerg 32:573–581
Wegrzyn JL, Eckert AJ, Choi M, Lee JM, Stanton BJ, Sykes R, Davis MF, Tsai CJ, Neale DB (2010) Association genetics of traits controlling lignin and cellulose biosynthesis in black cottonwood (Populus trichocarpa, Salicaceae) secondary xylem. New Phytol 188:515–532
Yang H, Yan R, Chen H, Zheng C, Lee DH, Liang DT (2006) In-depth investigation of biomass pyrolysis based on three major components: hemicellulose, cellulose, and lignin. Energ Fuel 20:388–393
Yang H, Yan R, Chen H, Lee DH, Zheng C (2007) Characteristics of hemicellulose, cellulose, and lignin pyrolysis. Fuel 86:1781–1788
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Zhong R, Morrison WH, Himmelsbach DS, Poole FL, Ye Z-H (2000) Essential role of caffeoyl coenzyme A O-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol 124:563–578
Acknowledgments
This work was funded by a grant from the USDA-CSREES McIntire-Stennis Cooperative Forestry Research Program to Dr. Larry B. Smart. Michelle Serapiglia was generously supported by the Josiah Lowe and Hugh Wilcox Scholarship Fund and the Edna Bailey Sussman Foundation. Appreciation is also expressed to Drs. William Powell, Larry Abrahamson, Tim Volk, Ed White, and Bill Winter for their support and advice as collaborators with this research and to Mark Appleby and Ken Burns for the excellent technical support. Special thanks go to Dr. Martin Goffinet for the assistance with histochemical and microscopy analysis of willow stems and to two anonymous reviewers for their insightful suggestions of improvements to this paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Brunner
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
PDF 389 kb
Rights and permissions
About this article
Cite this article
Serapiglia, M.J., Cameron, K.D., Stipanovic, A.J. et al. Correlations of expression of cell wall biosynthesis genes with variation in biomass composition in shrub willow (Salix spp.) biomass crops. Tree Genetics & Genomes 8, 775–788 (2012). https://doi.org/10.1007/s11295-011-0462-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-011-0462-7