Abstract
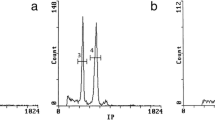
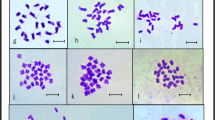
The subsection Asperae of genus Hydrangea L. (Hydrangeaceae) has been investigated for three reasons: several ambiguous classifications concerning Hydrangea aspera have been published, unexpected differences in genome size among seven accessions have been reported Cerbah et al. (Theor Appl Genet 103:45–51, 2001), and two atypical chromosome numbers (2n = 30 for Hydrangea involucrata and 2n = 34 for H. aspera) have been found when all other species of the genus present 2n = 36. Therefore, these two species and four subspecies of Hydrangea in all 29 accessions were analyzed for their genome size, chromosome number, and karyotype features. This investigation includes flow cytometric measurements of nuclear DNA content and bases composition (GC%), fluorochrome banding for detection of GC- and AT-rich DNA regions, and fluorescent in situ hybridisation (FISH) for chromosome mapping of 5 S and 18 S-5.8 S-26 S rDNA genes. In the H. aspera complex, the genome size ranged from 2.98 (subsp. sargentiana) to 4.67 pg/2C (subsp. aspera), an exceptional intraspecific variation of 1.57-fold. The mean base composition was 40.5% GC. Our report establishes the first karyotype for the species H. involucrata, and for the subspecies of H. aspera which indeed present different formulae, offering an element of discrimination. FISH and fluorochrome banding revealed the important differentiation between these two species (H. involucrata and H. aspera) and among four subspecies of the H. aspera complex. Our results are in agreement with the Chinese classification that places the groups Kawakami and Villosa as two different species: Hydrangea villosa Rehder and Hydrangea kawakami Hayata. This knowledge can contribute to effective germplasm management and horticultural use.



Similar content being viewed by others
References
Bennett MD, Bhandol P, Leitch IJ (2000) Nuclear DNA amounts in Angiosperms and their modern uses—807 new estimates. Ann Bot 86:859–909
Camacho JPM, Sharbel TF, Beukeboom LW (2000) B chromosome evolution. Philos Trans R Soc Lond, B 355:163–178
Castilho A, Heslop-Harrison JS (1995) Physical mapping of 5 S and 18 S–25 S rDNA and repetitive DNA sequences in Aegilops umbellata. Genome 38:91–96
Cerbah M, Coulaud J, Siljak-Yakovlev S (1998) rDNA organization and evolutionary relationships in the genus Hypochaeris (Asteraceae). J Heredity 89:312–318
Cerbah M, Mortreau E, Brown SC, Siljak-Yakovlev S, Bertrand H, Lambert C (2001) Genome size variation relationships in the genus Hydrangea. Theor Appl Genet 103:45–51
Chiche J, Brown SC, Leclerc JC, Siljak-Yakovlev S (2003) Genome size, heterochromatin organisation, and ribosomal gene mapping in four species of Ribes. Can J Bot 81:1049–1057
Chun WY (1954) A census and preliminary study of the Chinese Hydrangeoideae. Acta Phytotaxonomica Sinica 2:10–203
Cuéllar T, Orellana J, Belhassen E, Bella JL (1999) Chromosomal characterization and physical mapping of the 5 S and the 18 S–5.8 S–25 S ribosomal DNA in Helianthus argophyllus, with new data from Helianthus annuus. Genome 42:110–115
Danna KJ, Workman R, Coryell V, Keim P (1996) 5 S rRNA genes in tribe Phaseoleae: array size, number, and dynamics. Genome 39:445–455
De Melo NF, Guerra M (2003) Variability of the 5 S and 45 S rDNA Sites in Passiflora L. Species with distinct base chromosome numbers. Ann Bot 92:309–316
Doležel J, Greilhuber J, Suda J (2007) Flow cytometry with plant cells. Weinheim, Wiley-VCH Verlag
Funamoto T, Tanaka R (1988) Karyomorphological studies in some taxa of Hydrangea from Japan. Kromosomo 49:1583–1594
Furuta Y, Nishikawa K (1991) Variation in nuclear and individual chromosomal DNA contents and its role in evolution of plant. In: Gupta PK, Tsuchyia T (eds) Chromosome engineering in plants: genetics, breeding, evolution. Part A. Elsevier, New-York, pp 71–85
Geber G, Schweizer D (1987) Cytochemical heterochromatin differentiation in Sinapis alba (Cruciferae) using a simple air-drying technique for producing chromosome spreads. Plant Syst Evol 158:97–106
Gerlach WL, Dyer TA (1980) Sequence organization of the repeating units in the nucleus of wheat which contain 5 S rRNA genes. Nucleic Acids Res 8:4851–4865
Godelle B, Cartier D, Marie D, Brown CS, Siljak-Yakovlev S (1993) Heterochromatin study demonstrating the non-linearity of fluorometry useful for calculating genomic base composition. Cytometry 14:618–626
Guerra M (2000) Patterns of heterochromatin distribution in plant chromosomes. Genet Mol Biol 23:1029–1041
Hamon P, Siljak-Yakovlev S, Srisuwan S, Robin O, Poncet V, Hamon S, De Kochko A (2009) Physical mapping of rDNA and heterochomatin in 16 Coffea species: a revisited view of species differentiation. Chromosom res 17:291–304
Hasterok R, Jenkins G, Langdon TR, Jones N, Maluszynska J (2001) Ribosomal DNA is an effective marker of Brassica chromosomes. Theor Appl Genet 103:486–490
Heslop-Harrison JS, Schwarzacher T, Anamthawat-Jonsson K, Leitch AR, Shi M, Leitch IJ (1991) In situ hybridization with automated chromosome denaturation. Technique - A Journal of Methods in Cell and Molecular Biology 3:109–116
Hizume M, Ishida F, Murata M (1992) Multiple locations of the rRNA genes in chromosomes of pine Pinus densiflora and P. thumbergii. Jpn J Genet 67:389–396
Iwatsuki K, Boufford DE, Ohba H (2001) Flora of Japan, vol IIb. Kodansha, Japan, pp 84–94
Jones RN, Houben A (2003) B chromosomes in plants: escapees from the A chromosome genome? Trends Plant Sci 8:417–423
Jones RN, Diez M (2004) The B chromosome data base. Cytogenet Genome Res 106:149–150
Jones RN, Viegas W, Houben A (2008) A century of B chromosomes in plants: so what? Ann Bot 6:767–775
Kondo T, Hizume M (1982) Banding for the chromosomes of Cryptomeria japonica D. Don. J Jap For Soc 64:356–358
Lee SH, Do GS, Seo BB (1999) Chromosomal localization of 5 S rRNA gene loci and the implications for relationships within the Allium complex. Chrom Res 7:89–93
Leitch IJ, Heslop-Harrison JS (1992) Physical mapping of the 18 S–5.8 S–26 S rRNA genes in barley by in situ hybridization. Genome 35:1013–1018
Leitch IJ, Heslop-Harrison JS (1993) Physical mapping for four sites of 5 S rDNA sequences and one side of the alpha-amylase gene in barley (Hordeum vulgare). Genome 36:517–523
Levan A, Fredga D, Sandberg AA (1964) Nomenclature for centromeric positions on chromosomes. Hereditas 52:201–220
Levin DA, Palestris BG, Jones RN, Trivers R (2005) Phyletic hot spots for B chromosomes in angiosperms. Evolution 59:962–969
Maluszynska J, Heslop-Harrison JS (1993) Physical mapping of rDNA loci in Brassica species. Genome 36:774–781
Marie D, Brown SC (1993) A cytometric excercise in plant DNA histograms, with 2C values for 70 species. Biol Cell 78:41–51
McClintock E (1957) A monograph of the genus Hydrangea. Proc Calif Acad Sci 29:147–255
Mortreau E (2003) Etude de la variabilité génétique et de l'organisation génomique au sein d'une collection de ressources génétiques du genre Hydrangea. Ecole Nationale Supérieure agronomiques de Rennes, Ph D, p 150
Moscone EA, Klein F, Lambru M, Fuchs J, Schweizer D (1999) Quantitative karyotyping and dual-color FISH mapping of 5 S and 18 S–25 S rDNA probes in the cultivated Phaseolus species (Leguminosae). Genome 42:1224–1233
Murata M, Heslop-Harrison JS, Motoyoshi F (1997) Physical mapping of the 5 S ribosomal RNA genes in Arabidopsis thaliana by multi-color fluorescence in situ hybridization with cosmid clone. Plant J 12:31–37
Muratovic E, Bogunic F, Soljan D, Siljak-Yakovlev S (2005) Does Lilium bosniacum merit species rank? A classical and molecularcytogenetic approaches. Plant Syst Evol 252:97–109
Murray BG (2005) When does intraspecific C-value variation become taxonomically significant. Ann Bot 95:119–125
Ohri D (1998) Genome size variation and plant systematics. Ann Bot 82:75–83
Puertas MJ (2002) Nature and evolution of B chromosomes in plants: a non-coding but information-rich part of plant genome. Cytogenet Genome Res 96:198–205
Redher A (1911) Hydrangea. In: Sergent CS (ed) Plantae Wilsonianae. Cambridge University Press, Cambridge, pp 25–41
Saylor LC (1961) A karyotypic analysis of selected species of Pinus. Silvae Genet 10:77–84
Schmidt T, Schwarzacher T, Heslop-Harrison JS (1994) Physical mapping of rRNA genes by fluorescent in situ hybridization and structural analysis of 5 S rRNA genes and intergenic spacer sequences in sugar beet (Beta vulgaris). Theor Appl Genet 88:629–636
Schoennagel E (1931) Chromomenzahl und phylogenie der Saxifragaceen. Bot Jahrb 64:266–308
Schubert I, Wobus U (1985) In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 92:143–148
Schweizer D (1976) Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma 58:307–324
Shimizu T, Kao MT (1962) Saxifragaceae of Taiwan. Flora of Taiwan 8:127–142
Shimizu T, Kao MT (1977) Hydrangea L. In: Flora of Taiwan editorial committee, Taiwan, vol 3, pp 34–40
Siljak-Yakovlev S (1996) La dysploïdie et l'évolution du caryotype. Bocconea 5:210–220
Siljak-Yakovlev S, Cerbah M, Coulaud J, Stoian V, Brown SC, Zoldos V, Jelenic S, Papes D (2002) Nuclear DNA content, base composition, heterochromatin and rDNA in Picea omorika and Picea abies. Theor Appl Genet 104:505–512
Van Laere K, Van Huylenbroeck J, Van Bockstaele E (2007) Karyotype analysis and physical mapping of 45 S rRNA genes in Hydrangea species by fluorescence in situ hybridization. Plant Breed 127:301–307
Wei C, Bartholomew B (2001) Hydrangea. Flora of China. Beijing and Missouri Botanical Garden Press 8:145–157
Zoldos V, Papes D, Cerbah M, Panaud O, Besendorfer V, Siljak-Yakovlev S (1999) Molecular-cytogenetics studies of ribosomal genes organization among 11 Quercus species. Theor Appl Genet 99:969–977
Zonneveld BJM, Leicht IJ, Bennett MD (2006) First Nuclear DNA Amounts in more than 300 Angiosperms. Ann Bot 96:229–244
Acknowledgements
We thank Danièle Daury and Hervé Daniel from Institut National d'Horticulture (Angers) respectively for her technical support in cytogenetic and for his help in statistical analysis, Jean-Marc Bureau from the Institut des Sciences du Végétal (CNRS, Gif-sur-Yvette) and Odile Robin from Laboratoire Ecologie, Systématique et Evolution, UMR CNRS 8079 (Université Paris Sud, Orsay) for their expert assistance in flow cytometry and molecular cytogenetics on the IFR87 platform. This work was supported by the Région des Pays de la Loire.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Dirlewanger
Rights and permissions
About this article
Cite this article
Mortreau, E., Siljak-Yakovlev, S., Cerbah, M. et al. Cytogenetic characterization of Hydrangea involucrata Sieb. and H. aspera D. Don complex (Hydrangeaceae): genetic, evolutional, and taxonomic implications. Tree Genetics & Genomes 6, 137–148 (2010). https://doi.org/10.1007/s11295-009-0235-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-009-0235-8