Abstract
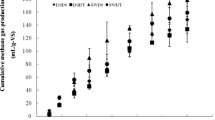
Mesophilic and thermophilic anaerobic digesters (MD and TD, respectively) utilizing Gracilaria and marine sediment as the substrate and inoculum, respectively, were compared by analyzing their performances and microbial community changes. During three successive transfers, the average cumulative methane yields in the MD and TD were 222.6 ± 17.3 mL CH4/g volatile solids (VS) and 246.1 ± 11 mL CH4/g VS, respectively. The higher hydrolysis rate and acidogenesis in the TD resulted in a several fold greater accumulation of volatile fatty acids (acetate, propionate, and butyrate) followed by a larger pH drop with a prolonged recovery than in the MD. However, the operational stability between both digesters remained comparable. Pyrosequencing analyses revealed that the MD had more complex microbial diversity indices and microbial community changes than the TD. Interestingly, Methanomassiliicoccales, the seventh methanogen order was the predominant archaeal order in the MD along with bacterial orders of Clostridiales, Bacteriodales, and Synergistales. Meanwhile, Coprothermobacter and Methanobacteriales dominated the bacterial and archaeal community in the TD, respectively. Although the methane yield is comparable, both MD and TD show a different profile of pH, VFA and the microbial communities.




Similar content being viewed by others
References
Adekunle KF, Okolie JA (2015) A review of biochemical process of anaerobic digestion. Adv Biosci Biotechnol 6:205–212
Akuzawa M, Hori T, Haruta S, Ueno Y, Ishii M, Igarashi Y (2011) Distinctive responses of metabolically active microbiota to acidification in a thermophilic anaerobic digester. Microbial Ecol 61:595–605
Amani T, Nosrati M, Sreekrishnan TR (2010) Anaerobic digestion from the viewpoint of microbiological, chemical, and operational aspects: a review. Environ Rev 18:255–278
APHA (1995) Standard methods for the examination of water and wastewater, 18th edn. American Public Health Association, Washington
Baena S, Fardeau ML, Labat M, Ollivier B, Garcia JL, Patel BKC (2000) Aminobacterium mobile sp. nov., a new anaerobic amino-acid-degrading bacterium. Int J Syst Evol Microbiol 50:259–264
Balch WE, Fox CE, Magrum LJ, Woese CR, Wolfe RS (1979) Methanogens: reevaluation of a unique biological group. Microbiol Rev 43:260–296
Borrel G, Harris HMB, Parisot N, Gaci N, Tottey W, Mihajlovski A, Deane J, Gribaldo S, Bardot O, Peyretaillade E, Peyret P, O’Toole PW, Brugère J-F (2013) Genome sequence of “Candidatus Methanomassiliicoccus intestinalis” Issoire-Mx1, a third Thermoplasmatales-related methanogenic archaeon from human feces. Genome Announc 1:e00453-13. doi:10.1128/genomeA.00453-13
Borrel G, Parisot N, Harris HMB, Peyretaillade E, Gaci N, Tottey W, Bardot O, Raymann K, Gribaldo S, Peyret P, O’Toole PW, Brugère JF (2014) Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics BioMed Central 15(1):679
Cai S, Dong X (2010) Cellulosilyticum ruminicola gen. nov., sp. nov., isolated from the rumen of yak, and reclassification of Clostridium lentocellum as Cellulosilyticum lentocellum comb. nov. Int J Syst Evol Microbiol 60:845–849
Cantrell KB, Ducey T, Ro KS, Hunt PG (2008) Livestock waste–to- bioenergy generation opportunities. Bioresour Technol 99:7941–7953
Chen M (1983) Adaptation of mesophilic anaerobic sewage fermentor populations to thermophilic temperatures. Appl Environ Microbiol 45:1271–1276
Chynoweth DP, Owens JM, Legrand R (2001) Renewable methane from anaerobic digestion of biomass. Renew Energy 22:1–8
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci USA 89:5685–5689
Desvaux M (2005) Clostridium cellulolyticum: model organism of mesophilic cellulolytic clostridia. FEMS Microbiol Rev 29(4):741–764
Eddy SR (2011) Accelerated profile HMM searches. PLoS Comput Biol 7(10):e1002195
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200
Felsenstein J (1985) Phylogenies and the comparative method. Am Nat 125:1–15
Franke-Whittle IH, Walter A, Ebner C, Insam H (2014) Investigation into the effect of high concentrations of volatile fatty acids in anaerobic digestion on methanogenic communities. Waste Manag 34(11):2080–2089
Garcia LS, Jangid K, Whitman WB, Das KC (2011) Transition of microbial communities during the adaption to anaerobic digestion of carrot waste. Bioresour Technol 102:7249–7256
Green RH (1989) Power analysis and practical strategies for environmental monitoring. Environ Res 50:195–205
Hattori S, Kamagata Y, Hanada S, Shoun H (2000) Thermacetogeniumphaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate-oxidizing bacterium. Int J Syst Evol Microbiol 50:1601–1609
Honda T, Fujita T, Tonouchi A (2013) Aminivibrio pyruvatiphilus gen. nov., sp. nov., an anaerobic, amino-acid-degrading bacterium from soil of a Japanese rice field. Int J Syst Evol Microbiol 63:3679–3686
Hughes AD, Kelly MS, Black KD, Stanley MS (2012) Biogas from macroalgae: is it time to revisit the idea? Biotechnol Biofuels 5:86
Jeon YS, Chun J, Kim BS (2013) Identification of household bacterial community and analysis of species shared with human microbiome. Curr Microbiol 67(5):557–563
Jukes TH, Cantor CR (1969) Evolution of protein molecules. Academic Press, New York, pp 21–132
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Li WW, Yu HQ (2011) Biohydrogen production with high-rate bioreactors. In: Pandey A et al (eds) Biofuels alternative feedstocks and conversion processes. Elsevier, San Diego
Li YF, Nelson MC, Chen PH, Graf J, Li Y, Yu Z (2015) Comparison of the microbial communities in solid-state anaerobic digestion (SS-AD) reactors operated at mesophilic and thermophilic temperatures. Appl Microbiol Biotechnol 99(2):969–980
Liu Y, Qiao JT, Yuan XZ, Guo RB, Qiu YL (2014) Hydrogenispora ethanolica gen. nov., sp. nov., an anaerobic carbohydrate-fermenting bacterium from anaerobic sludge. Int J Syst Evol Microbiol 64:1756–1762
Mhuantong W, Charoensawan V, Kanokratana P, Tangphatsornruang S, Champreda V (2015) Comparative analysis of sugarcane bagasse metagenome reveals unique and conserved biomass-degrading enzymes among lignocellulolytic microbial communities. Biotechnol Biofuels 8:16
Migliore G, Alisi C, Sprocati AR, Massi E, Ciccoli R, Lenzi M, Wang A, Cremisini C (2012) Anaerobic digestion of macroalgal biomass and sediments sourced from the Orbetello lagoon, Italy. Biomass Bioenergy 42:69–77
Miura T, Kita A, Okamura Y, Aki T, Matsumura Y, Tajima T, Kato J, Nakashimada Y (2014) Evaluation of marine sediments as microbial sources for methane production from brown algae under high salinity. Bioresour Technol 169:362–366
Nesbø CL, Bradnan DM, Adebusuyi A, Dlutek M, Petrus AK, Foght J, Doolittle WF, Noll KM (2012) Mesotoga prima gen. nov., spnov., the first described mesophilic species of the Thermotogales. Extremophiles 16:387–393
Oren A, Garrity GM (2013) List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 63:3931–3934
Pelletier E, Kreimeyer A, Bocs S, Rouy Z, Gyapay G, Chouari R, Rivière D, Ganesan A, Daegelen P, Sghir A (2008) ‘‘Candidatus Cloacamonas acidaminovorans’’: genome sequence reconstruction provides a first glimpse of a new bacterial division. J Bacteriol 190(7):2572–2579
Pope PB, Vivekanand V, Eijsink VGH, Horn SJ (2013) Microbial community structure in a biogas digester utilizing the marine energy crop Saccharina latissimi. Biotech 3:407–414
Rode LM, Genthner BR, Bryant MP (1981) Syntrophic association by cocultures of the methanol- and CO(2)-H(2)-utilizing species Eubacterium limosum and pectin-fermenting Lachnospiramultiparus during growth in a pectin medium. Appl Environ Microbiol 42:20–22
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Sasaki K, Morita M, Sasaki D, Nagaoka J, Matsumoto N, Ohmura N, Shinozaki H (2011) Syntrophic degradation of proteinaceous materials by the thermophilic strains Coprothermobacter proteolyticus and Methanothermobacter thermautotrophicus. J Biosci Bioeng 112:469–472
Schink B, Zeikus J (1980) Microbial methanol formation: a major end product of pectin metabolism. Curr Microbiol 4:387–389
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541
Schramm W, Lehnberg W (1984) Mass culture of brackish-water-adapted seaweeds in sewage-enriched seawater II. Fermentation for biogas production. Hydrobiologia 116–117(1):276–281
Shi J, Wang Z, Stiverson JA, Yu Z, Li Y (2013) Reactor performance and microbial community dynamics during solid-state anaerobic digestion of corn stover at mesophilic and thermophilic conditions. Bioresour Technol 136:574–581
Snelling TJ, Genc B, McKain N, Watson M, Waters SM, Creevey CJ, Wallace RJ (2014) Diversity and community composition of methanogenic archaea in the rumen of Scottish upland sheep assessed by different methods. PLoS ONE 9(9):e106491
Su XL, Tian Q, Zhang J, Yuan XZ, Shi XS, Guo RB, Qiu YL (2014) Acetobacteroides hydrogenigenes gen. nov., sp. nov., an anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int J Syst Evol Microbiol 64(9):2986–2991
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Vanegas CH, Bartlett J (2012) Anaerobic digestion of Laminaria digitata: the effect of temperature on biogas production and composition. Waste Biomass Valor 4:509–515. doi:10.1007/s12649-012-9181-z
Vanegas CH, Bartlett J (2013) Green energy from marine algae: biogas production and composition from the anaerobic digestion of Irish seaweed species. Environ Technol 34(15):2277–2283
Wang Y, Zhang Y, Wang J, Meng L (2009) Effects of volatile fatty acid concentrations on methane yield and methanogenic bacteria. Biomass Bioenergy 33(5):848–885
Wang X, Sharp CE, Jones GM, Grasby SE, Brady AL, Dunfield PF (2015) Stable-isotope-probing identifies uncultured Planctomycetes as primary degraders of a complex heteropolysaccharide in soil. Appl Environ Microbiol 81:4607–4615
Wei N, Quarterman J, Jin YS (2013) Marine macroalgae: an untapped resource for producing fuels and chemicals. Trends Biotechnol 31:70–77
Westerholm M, Roosand S, Schnürer A (2011) Tepidanaerobacter acetatoxydans sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated from two ammonium-enriched mesophilic methanogenic processes. Syst Appl Microbiol 34:260–266
Yokoyama H, Wagner ID, Wiegel J (2010) Caldicoprobacter oshimai gen. nov., sp. nov., an anaerobic, xylanolytic, extremely thermophilic bacterium isolated from sheep faeces, and proposal of Caldicoprobacteraceae fam. nov. Int J Syst Evol Microbiol 60:67–71
Acknowledgments
This study was supported by the Marine Biotechnology Research Division, Korea Institute of Ocean Science and Technology (KIOST) in-house program (PE99413) and C1 Gas Refinery Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning in the Republic of Korea (2015M3D3A1A01064884).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict interests
The authors have declared that no competing interests exist.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Azizi, A., Kim, W. & Lee, J.H. Comparison of microbial communities during the anaerobic digestion of Gracilaria under mesophilic and thermophilic conditions. World J Microbiol Biotechnol 32, 158 (2016). https://doi.org/10.1007/s11274-016-2112-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-016-2112-6