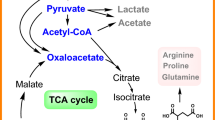
Abstract
The regulation of metabolic flux through glycolytic versus the gluconeogenic pathway plays an important role in central carbon metabolism. In this study, we made an attempt to enhance riboflavin production by deregulating gluconeogenesis in Bacillus subtilis. To this end, gapB (code for NADPH-dependent glyceraldehyde-3-phosphate dehydrogenase), fbp (code for fructose-1,6-bisphosphatase) and pckA (code for phosphoenolpyruvate carboxykinase) were overexpressed in parental strain B. subtilis RH33. Compared with RH33, overexpression of fbp and gapB resulted in approximately 18.0 and 14.2 % increased riboflavin production, respectively, while overexpression of pckA obtained the opposite result. Significant enhancement of riboflavin titers up to 4.89 g/l was obtained in shake flask cultures when gapB and fbp were co-overexpressed, nevertheless the specific growth rate decreased slightly and the specific glucose uptake rate remained almost unchanged. An improvement by 21.9 and 27.8 % of the riboflavin production was achieved by co-overexpression of gapB and fbp in shake flask and fed-batch fermentation, respectively. These results imply that deregulation of gluconeogenesis is an effective strategy for production of metabolites directly stemming from the pentose phosphate pathway as well as other NADPH-demanding compounds with glucose as carbon source in B. subtilis.




Similar content being viewed by others
Abbreviations
- PPP:
-
Pentose phosphate pathway
- Ru5P:
-
Ribulose-5P
- FBPase:
-
Fructose 1,6-bisphosphatase
- MM:
-
Minimal medium
- CDW:
-
Cell dry weight
- TCA:
-
Tricarboxylic acid
- PEP:
-
Phosphoenolpyruvate
References
Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, Sekowska A, Vallenet D, Wang T, Moszer I, Medigue C, Danchin A (2009) From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155(Pt 6):1758–1775
Becker J, Klopprogge C, Zelder O, Heinzle E, Wittmann C (2005) Amplified expression of fructose 1,6-bisphosphatase in Corynebacterium glutamicum increases in vivo flux through the pentose phosphate pathway and lysine production on different carbon sources. Appl Environ Microbiol 71(12):8587–8596
Becker J, Klopprogge C, Herold A, Zelder O, Bolten CJ, Wittmann C (2007) Metabolic flux engineering of l-lysine production in Corynebacterium glutamicum-over expression and modification of G6P dehydrogenase. J Biotechnol 132(2):99–109
Blencke HM, Homuth G, Ludwig H, Mader U, Hecker M, Stulke J (2003) Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab Eng 5(2):133–149
Coquard D, Huecas M, Ott M, van Dijl JM, van Loon AP, Hohmann HP (1997) Molecular cloning and characterisation of the ribC gene from Bacillus subtilis: a point mutation in ribC results in riboflavin overproduction. Mol Gen Genet 254(1):81–84
Dauner M, Bailey JE, Sauer U (2001a) Metabolic flux analysis with a comprehensive isotopomer model in Bacillus subtilis. Biotechnol Bioeng 76(2):144–156
Dauner M, Storni T, Sauer U (2001b) Bacillus subtilis metabolism and energetics in carbon-limited and excess-carbon chemostat culture. J Bacteriol 183(24):7308–7317
Dauner M, Sonderegger M, Hochuli M, Szyperski T, Wuthrich K, Hohmann HP, Sauer U, Bailey JE (2002) Intracellular carbon fluxes in riboflavin-producing Bacillus subtilis during growth on two-carbon substrate mixtures. Appl Environ Microbiol 68(4):1760–1771
Duan YX, Chen T, Chen X, Zhao XM (2010) Overexpression of glucose-6-phosphate dehydrogenase enhances riboflavin production in Bacillus subtilis. Appl Microbiol Biotechnol 85(6):1907–1914
Fillinger S, Boschi-Muller S, Azza S, Dervyn E, Branlant G, Aymerich S (2000) Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J Biol Chem 275(19):14031–14037
Fischer M, Haase I, Kis K, Meining W, Ladenstein R, Cushman M, Schramek N, Huber R, Bacher A (2003) Enzyme catalysis via control of activation entropy: site-directed mutagenesis of 6,7-dimethyl-8-ribityllumazine synthase. J Mol Biol 326(3):783–793
Fujita Y, Yoshida K, Miwa Y, Yanai N, Nagakawa E, Kasahara Y (1998) Identification and expression of the Bacillus subtilis fructose-1, 6-bisphosphatase gene (fbp). J Bacteriol 180(16):4309–4313
Hemberger S, Pedrolli DB, Stolz J, Vogl C, Lehmann M, Mack M (2011) RibM from Streptomyces davawensis is a riboflavin/roseoflavin transporter and may be useful for the optimization of riboflavin production strains. BMC Biotechnol 11:119
Humbelin M, Griesser V, Keller T, Schurter W, Haiker M, Hohmann HP, Ritz H, Richter G, Bacher A, van Loon APGM (1999) GTP cyclohydrolase II and 3,4-dihydroxy-2-butanone 4-phosphate synthase are rate-limiting enzymes in riboflavin synthesis of an industrial Bacillus subtilis strain used for riboflavin production. J Ind Microbiol Biotechnol 22(1):1–7
Jules M, Le Chat L, Aymerich S, Le Coq D (2009) The Bacillus subtilis ywjI (glpX) gene encodes a class II fructose-1,6-bisphosphatase, functionally equivalent to the class III Fbp enzyme. J Bacteriol 191(9):3168–3171
Kalingan AE, Liao CM (2002) Influence of type and concentration of flavinogenic factors on production of riboflavin by Eremothecium ashbyii NRRL 1363. Bioresour Technol 82(3):219–224
Lehmann M, Degen S, Hohmann HP, Wyss M, Bacher A, Schramek N (2009) Biosynthesis of riboflavin: screening for an improved GTP cyclohydrolase II mutant. FEBS J 276(15):4119–4129
Li XJ, Chen T, Chen X, Zhao XM (2006) Redirection electron flow to high coupling efficiency of terminal oxidase to enhance riboflavin biosynthesis. Appl Microbiol Biotechnol 73(2):374–383
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408
Perkins JB, Sloma A, Hermann T, Theriault K, Zachgo E, Erdenberger T, Hannett N, Chatterjee NP, Williams V, Rufo GA, Hatch R, Pero J (1999) Genetic engineering of Bacillus subtilis for the commercial production of riboflavin. J Ind Microbiol Biotechnol 22(1):8–18
Ruhl M, Zamboni N, Sauer U (2010) Dynamic flux responses in riboflavin overproducing Bacillus subtilis to increasing glucose limitation in fed-batch culture. Biotechnol Bioeng 105(4):795–804
Sauer U, Eikmanns BJ (2005) The PEP–pyruvate–oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev 29(4):765–794
Sauer U, Hatzimanikatis V, Hohmann HP, Manneberg M, van Loon AP, Bailey JE (1996) Physiology and metabolic fluxes of wild-type and riboflavin-producing Bacillus subtilis. Appl Environ Microbiol 62(10):3687–3696
Sauer U, Cameron DC, Bailey JE (1998) Metabolic capacity of Bacillus subtilis for the production of purine nucleosides, riboflavin, and folic acid. Biotechnol Bioeng 59(2):227–238
Schallmey M, Singh A, Ward OP (2004) Developments in the use of Bacillus species for industrial production. Can J Microbiol 50(1):1–17
Servant P, Le Coq D, Aymerich S (2005) CcpN (YqzB), a novel regulator for CcpA-independent catabolite repression of Bacillus subtilis gluconeogenic genes. Mol Microbiol 55(5):1435–1451
Shi S, Chen T, Zhang Z, Chen X, Zhao X (2009a) Transcriptome analysis guided metabolic engineering of Bacillus subtilis for riboflavin production. Metab Eng 11(4–5):243–252
Shi SB, Shen Z, Chen X, Chen T, Zhao XM (2009b) Increased production of riboflavin by metabolic engineering of the purine pathway in Bacillus subtilis. Biochem Eng J 46(1):28–33
Sklyarova SA, Kreneva RA, Perumov DA, Mironov AS (2012) The characterization of internal promoters in the Bacillus subtilis riboflavin biosynthesis operon. Russ J Genet 48(10):967–974
Stahmann KP, Revuelta JL, Seulberger H (2000) Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production. Appl Microbiol Biotechnol 53(5):509–516
Tannler S, Fischer E, Le Coq D, Doan T, Jamet E, Sauer U, Aymerich S (2008a) CcpN controls central carbon fluxes in Bacillus subtilis. J Bacteriol 190(18):6178–6187
Tannler S, Zamboni N, Kiraly C, Aymerich S, Sauer U (2008b) Screening of Bacillus subtilis transposon mutants with altered riboflavin production. Metab Eng 10(5):216–226
Wang Z, Chen T, Ma X, Shen Z, Zhao X (2011) Enhancement of riboflavin production with Bacillus subtilis by expression and site-directed mutagenesis of zwf and gnd gene from Corynebacterium glutamicum. Bioresour Technol 102(4):3934–3940
Wu QL, Chen T, Gan Y, Chen X, Zhao XM (2007) Optimization of riboflavin production by recombinant Bacillus subtilis RH44 using statistical designs. Appl Microbiol Biotechnol 76(4):783–794
Yoshida K, Kobayashi K, Miwa Y, Kang CM, Matsunaga M, Yamaguchi H, Tojo S, Yamamoto M, Nishi R, Ogasawara N, Nakayama T, Fujita Y (2001) Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res 29(3):683–692
Zamboni N, Mouncey N, Hohmann H-P, Sauer U (2003) Reducing maintenance metabolism by metabolic engineering of respiration improves riboflavin production by Bacillus subtilis. Metab Eng 5(1):49–55
Zhu Y, Chen X, Chen T, Zhao X (2007) Enhancement of riboflavin production by overexpression of acetolactate synthase in a pta mutant of Bacillus subtilis. FEMS Microbiol Lett 266(2):224–230
Acknowledgments
We thank Dr. Zhenquan Lin for the help of qRT-PCR analysis. This work was supported by National Program on Key Basic Research Project (2011CBA00804, 2012CB725203), National Natural Science Foundation of China (NSFC-21206112, NSFC-21176182), National High-tech R&D Program of China (2012AA022103, 2012AA02A702) and the Innovation Foundation of Tianjin University (1308).
Author information
Authors and Affiliations
Corresponding author
Additional information
Guanglu Wang and Ling Bai have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, G., Bai, L., Wang, Z. et al. Enhancement of riboflavin production by deregulating gluconeogenesis in Bacillus subtilis . World J Microbiol Biotechnol 30, 1893–1900 (2014). https://doi.org/10.1007/s11274-014-1611-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-014-1611-6