Abstract
In this study, the genome sequence of a norovirus GII.4 strain isolated from South China was comparatively analyzed. The RNA genome of the strain GZ2013-L10 was composed of 7513 nucleotides. Phylogenetic analyses based on three ORFs confirmed its genotype as GII.Pe/GII.4-2012. Compared with other 22 genomes of the same variant, nine distinct nucleotide substitutions were found in the new genome, which resulted in three amino acid changes. All 138 capsid protein VP1 sequences of GII.4-2012 variants were also collected, and multiple alignments revealed 35 variable codons. Evolutionary analyses of GII.4-2012 variants were performed against previous pandemic GII.4 variants, and 2 distinctive changes were identified on epitopes A and E (E368, T413), which resulted in an obvious variation of their solvent-accessible surface areas. Therefore, the genome of GZ2013-L10 was extensively characterized, and new emerging variations on viral epitopes were predicted to contribute to NoV persistence in humans.
Similar content being viewed by others
Introduction
Norovirus (NoV) is the major cause of acute non-bacterial diarrhea worldwide [1, 2]. The virus belongs to the family Caliciviridae, and it has a positive-sense single-stranded RNA genome of 7.5–7.7 kb in size, which consists of three open reading frames (ORFs). Specifically, ORF1 encodes a polyprotein that could be cleaved into six non-structural proteins (NS1/2-NS7), and remaining two ORFs encode a major structural protein VP1 and an alkaline minor structural protein VP2, respectively [3]. The viral capsid contains 180 major capsid protein VP1 monomers, which is composed of a shell domain (S) and a protruding arm (P) with a flexible hinge. The P arm could be subdivided into P1-1, P2, and P1-2 subregions, and P2 is associated with the receptor-binding function and viral immunogenicity [4, 5].
As an RNA virus, NoV has a rich genetic diversity. Based on the capsid VP1 amino acid sequences, NoVs can be classified into six genogroups (GI–GVI), and a seventh genogroup has also been proposed recently [6, 7]. Additionally, most genogroups could be further subdivided into different genotypes, with over 40 described up to now [8]. However, only GII.4 variant is the predominant genotype that caused majority of the outbreaks and sporadic infections worldwide [2]. To date, GII.4 genotype contained about 14 epidemic variants according to the genetic diversity of the P2 subregion.
The severity of GII.4 NoV has been studied comprehensively since its global epidemic. New GII.4 variants frequently emerged and clearly replaced the former dominant variant every 2 or 3 years. The ancestral GII.4 variant was collected in 1974, but it did not cause global pandemic until the appearance of the US 95/96 variant in the mid-1990s [9, 10]. Since then, a series of GII.4 variants have been found worldwide and became the predominant genotypes, including GII.4-2002 (Farmington variant), GII.4-2004 (Hunter variant), GII.4-2006a (Yerseke variant), GII.4-2006b (Den Haag variant), GII.4-2007EU (Apeldoorn variant), GII.4-2009 (New Orleans variant), and GII.4-2012 (Sydney variant). The last variant GII.4-2012 was first identified in March 2012 in Australia [11], and then the emergence of the new variant caused an obvious increase of NoV-related outbreaks and sporadic infections in many countries, including China [12–15].
Data of viral sequences provided an essential basis for NoV researches, and the importance of viral genome sequencing has recently drawn more attention worldwide [16–18]. Since recombination was presumed as one of the major evolutionary mechanisms of these viruses [19], it was necessary to analyze the entire genome sequence, which would aid in the accurate evaluation of the epidemiological feature as well as the discovery of new antigenic types. Therefore, in this study, the complete nucleotide sequence of GZ2013-L10 strain detected in South China was identified and comparatively analyzed with other related NoV variants circulating worldwide.
Materials and methods
Sample collection and genome sequencing
An NoV strain GZ2013-L10 was detected during an NoV surveillance study in South China. Sequencing of the NoV genome was performed according to the previous protocol [20]. In brief, viral RNA was first extracted from the fecal supernatant and then used as a template for the NoV genome amplification. All six fragments encompassing the viral genome were generated via one-step reverse transcription PCR (TAKARA, Dalian, China). The RT-PCR products were then sequenced directly using an automated sequencer (ABI 3730XL, Applied Biosystems, Foster City, CA, USA), and the viral genome was constructed by connecting overlapped sections as a single sequence.
Sequence analyses
Similarity searches of GZ2013-L10 strain were performed using the BLAST search algorithm, and ORF positions were verified using the ORF finder and compared with representative sequences (accession no. JX459908). Multiple alignments were performed using ClustalX for Windows (version 1.83) with the default parameters [21]. The phylogenetic relationship of GZ2013-L10 and the representative NoV strains from each genogroup were assessed using the software MEGA version 6.0 [22]. The reliability of the different phylogenetic groupings was evaluated by bootstrap test (1000 replications). All nucleotide sequences were also genotyped using the NoV automated online genotyping tool (www.rivm.nl/mpf/norovirus/typingtool) [23]. The capsid protein VP1 sequences of GII.4-2012 NoV strains were compared to investigate differences at the amino acid level using BioEdit version 7.0.1.
Homology modeling
Homology modeling was performed through the Automated Comparative Protein Modeling Server of the Swiss Institute of Bioinformatics [24]. Changes to the solvent-accessible surface areas of variable sites on modeled structures of viral capsid proteins VP1 were first calculated (in Å2) using the SASA command provided in VMD [25], and these structures were also submitted to the protein antigen spatial epitope prediction web server (SEPPA) for predicting possible spatial epitopes with the default threshold [26]. Protein structures were visualized and manipulated using PyMOL v1.4.1 (DeLano Scientific, LLC, San Francisco, CA).
Reference NoV strains
Reference NoV strains with entire genomes used in this study are listed in Table 1.
Results
Genome composition of the GZ2013-L10 strain
During an NoV surveillance study in South China in the winter of 2013, a fecal sample GZ2013-L10 from a 7-month-old male infant with acute diarrhea was identified as GII NoV positive. The strain GZ2013-L10 (accession no. KT202793) showed the genome length of 7513 nucleotides with three complete ORFs, which were spanning nucleotides 5–5104, 5085–6707, and 6707–7513, respectively. BLAST results with the newly obtained genome nucleotide sequence as the search query showed high similarity with 22 NoV sequences (query coverage ≥99 % and maximum identity ≥98 %), which were isolated from Australia, Bangladesh, Canada, the Chinese mainland, Japan, the Hong Kong region, and the Taiwan region over the last 3 years, respectively.
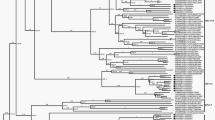
Based on multiple alignments, the genetic relationship of GZ2013-L10 with reference NoV strains of different genotypes with entire genomes was analyzed. And the result revealed the inconsistency of pairwise identities for different ORFs. With respect to ORF1, the nucleotide sequence and amino acid sequence identities of GZ2013-L10 with reference human NoV strains (GI, GII, and GIV genogroups) ranged from 55.7 to 99.1 and from 53.6 to 99.4 %, respectively. In the case of ORF2, the nucleotide sequence and amino acid sequence identities ranged from 50.4 to 99.0 and from 43.9 to 99.3 %, respectively. When compared for ORF3, the nucleotide sequence and amino acid sequence identities ranged from 40.1 to 98.5 and from 36.9 to 97.4 %, respectively. Specifically, the GZ2013-L10 strain was most closely related to the GII.4-2012 strain for each ORF at both nucleotide and amino acid levels. The genotype of GZ2013-L10 strain was identified as GII.4-2012 by submitting its genome sequence to NoV online genotyping tool, and the result was identical to those of phylogenetic analyses based on each ORF at the nucleotide level (Fig. 1a–c).
Phylogenetic analyses based on nucleotide sequences of three ORFs of representative strains for each NoV genotype: a ORF1, b ORF2, and c ORF3. A neighbor joining method was used based on the Kimura two-parameter distance model. Bootstrap analysis was done using 1000 replicates and the results were expressed as percentages at the nodes. The scale bar represents the expected number of substitutions per site. GZ2013-L10 is highlighted with a black triangle. GZ Guangzhou
Comparative analyses with other GII.4-2012 genomes
To verify the variation of GII.4-2012 NoV during its global epidemics, the genomic differences of the strain GZ2013-L10 from South China were compared with other 22 reported GII.4-2012 genomes. Among these strains, 8 were isolated from the Taiwan region, 4 from Japan, 3 from the Hong Kong region, 2 from Australia, 2 from Bangladesh, 2 from the Chinese mainland, and 1 from Canada between 2008 and 2012. The pairwise identity of GZ2013-L10 with other NoV GII.4-2012 strains was also analyzed for three ORFs, and the results revealed high sequence similarities at the nucleotide (98.4–99.7, 97.5–99.8, 97.6–99.6 %) and amino acid levels (98.9–99.8, 98.3–99.8, 97.0–99.3 %), respectively. Based on multiple alignments, disagreements among the genome sequence of 23 GII.4-2012 NoV strains were 390nt/104aa in ORF1, 114nt/27aa in ORF2, and 64nt/28aa in ORF3, respectively. However, only 9 distinct nucleotide substitutions (C240, A245, T1171, C1588, G3153, C4201, A5273, T6095, T6365) occurred with the emergence of GZ2013-L10 strain, which resulted in 3 amino acid changes in ORF1 (P79, I81, C389).
Sequence analysis of the capsid protein VP1 of GII.4-2012
To further explore the diversity of the capsid protein VP1 of the GII.4-2012 variant at the amino acid level, another 115 capsid protein VP1 sequences of the same genotype were also collected and analyzed along with the former representative strains. The capsid protein VP1 of GZ2013-L10 was found identity to those of six strains isolated in Australia (KF060020, KF060027, KF060033, KF060042, and KF060067) and New Zealand (KF060128). And there were also eight strains which showed high pairwise identity (99.8 %), which were from Japan (KJ196279, KJ196280, KJ196281, KJ196293), the Hong Kong region (JX629458, KC175323), and Australia (KF060046, KF060073), respectively. On the contrary, the strains in New Zealand (KF060122 and KF060123) showed low pairwise identity (97.0 %). Meanwhile, there were 8 strains isolated from the Chinese mainland, which showed pairwise identity with the new strain between 98.0 and 99.6 %. Furthermore, on the basis of multiple alignments, there were 35 variable sites on the capsid protein VP1 of GII.4-2012 variants, and 19 of these variations were located in the P2 subregion (Fig. 2). Specially, ten variable sites showed high variability, where only <90 % strains employed the same amino acid, including V119, V145, P174, N309, V333, H373, G393 H414, A539, and V540.
Variation sites in the capsid protein VP1 of GII.4-2012 variant. Sites in P2 region are highlighted in purple above the amino acids, with the N-terminal and C-terminal flanking regions of heterogeneity in green for the S domain and blue for P1. Ten variable sites with high variability are highlighted in yellow (Color figure online)
Over the last two decades, the epidemiological pattern of NoV GII.4 variants has represented an obvious epochal evolution process, which is a novel variant emerged every 2–3 years to replace its predecessors. Therefore, the capsid protein VP1 of other previous GII.4 pandemic variants was also collected for evolutionary analyses with the novel GII.4-2012 variant. Focused on five blockage epitopes A–E, only 2 sites were to be found specific to the new variant, which were located in epitopes A and E (E368 and T413), respectively. Besides, the usage of amino acid on the other sites was inherited from its successive GII.4 variant lineages, including GII.4-2009 (Prototype: GU445325| New Orleans1805), GII.4-2007EU (Prototype: AB445395|Apeldoorn317), GII.4-2007JP (Prototype: AB541319|Osaka1), and GII.4-2006b (Prototype: EF126965|DenHaag89) (Fig. 3). And then, the effect of these variations on the capsid protein VP1 of GII.4-2012 was evaluated by homology modeling, with the published crystal structure of GII.4 NoV strain VA387 (PDB: 2OBT) as a template. By submitting to SEPPA to predict possible spatial epitopes, the new emerging amino acid E368 (1.77) and T413 (1.99) also showed high antigenicity scores as the former amino acid A368 (1.79) and I413 (1.99), which indicated high antigenicity of the new variant. Meanwhile, variations of solvent-accessible surface areas of these sites were analyzed. As a result, it was found that the same amino acid used by different variants at the same position often had the similar surface areas, such as T294:133.5, S296:41.6, R297:110.7, N298:26.1, V333:45.2, K382:58.7, T340:117.7, E376:44.6, G393:80.0, T394:83.9, T395:84.4, S407:46.2, and N412:121.8. And the usage of the two new amino acids caused an obvious variation of surface areas (A368E: from 55.8 to 102.2, I413T: from 94.2 to 73.0). Besides, a neighboring conserved residue D372 was affected reducing its surface area (from 53.7 to 40.7).
Evolution sites in the epitopes A–E of GII.4-2012 variant based on the alignment with other GII.4 strains. GII.4-2012 consensus means the consensus sequence of GII.4-2012 variant, which reflected the majority of the amino acid sequences of the 138 GII.4-2012 NoV strains. The numbers under the amino acids indicated its solvent-accessible surface area and antigenicity score. Two distinct residues (368 and 413) of GII.4-2012 are highlighted in light gray
Discussion
NoVs are regarded as the leading viral agent of epidemic and sporadic gastroenteritis in most countries, including China [2]. As an RNA virus, rich genetic diversity is one of the primary obstacles to successful NoV vaccination, since minor variation on the capsid protein VP1 could affect viral immunogenicity. Recently, more and more complete genomes were accumulated for understanding NoV genetic diversity [18]. In this study, the genome sequence of the GZ2013-L10 strain detected in South China was identified and comparatively analyzed.
Among over 40 NoV genotypes reported, GII.4 is the main variant for most related outbreaks and infections. And a series of GII.4 variants have become the most predominant strains since their first global epidemic in the mid-1990s, with a 2- or 3-year interval. Based on phylogenetic analyses with reference NoV strains of different genotypes, the GZ2013-L10 strain detected in South China was found to belong to GII.4-2012 variant. The novel GII.4 variant was first detected in Australia [11], and then it was reported as the predominant genotype which caused an obvious increase of NoV activity levels compared to the previous seasons in most countries, including the US, Spain, Italy, and China [13, 27–29].Therefore, it was necessary and urgent to further understand GII.4-2012 variant due to its severity.
It is known that the RNA genome of NoV consists of three ORFs, but the homology of different ORFs is not consistent with each other. When the sequences of each ORF of GZ2013-L10 strain were compared with those of NoV strains of different genotypes, the ORF1 region was shown as the most conserved one with high pairwise identities at both nucleotide and amino acid levels, followed by ORF2 and ORF3. The similarity of GZ2013-L10 with the strains belonging to the same GII.4-2012 variant was also analyzed, the results of which revealed the inconsistency of three ORFs as above. In the past studies, it was found that p48 (NS1-2) and p22 in the ORF1 were the variable regions as VP1 and VP2 [18]. In spite of the lack of understanding of these proteins except VP1, it was predicted that these variable regions among the viral genomes were of significance for the persistence of NoV in humans, which should be further explored in the future.
The evolution of viral capsid protein VP1 was regarded as being of great significance for NoV prevalence, which was mainly due to its distribution of receptor-binding sites and antigenic epitopes [5, 30]. Therefore, all VP1 sequences of GII.4-2012 variant were collected which were mainly detected in Australia, New Zealand, Japan, the Chinese mainland, the Hong Kong region, the Taiwan region, France, Italy, Canada, and Bangladesh. However, the results of homology analyses of GZ2013-L10 did not show a regional correlation that it was closely related to the strains detected in Australia and New Zealand rather than those in the Chinese mainland. It might be due to the rapid spreading of NoVs, and some studies reported that it could spread all over the world in 3 months [31]. So it pointed out the necessity of establishments of global NoV surveillance networks as an approach to guide preventive measures against new GII.4 variant emerged.
Based on the crystal structure, viral capsid protein VP1 could be divided into different subregions, of which the outermost P2 was the hypervariable portion. When comparative analyses were performed with the GII.4-2012 variant, more than half (19/35) of the variable sites were found to be located in the P2 subregion and therefore half of the ten variable sites with high variability. Moreover, herd immunity and host receptor recognition were regarded as the major factors driving the emergence of novel GII.4 variants, which was also mainly embodied in the P2 subregion [5, 30]. Some studies indicated that the GII.4 NoV was continuously evolving through the alteration of the surface-exposed receptor-binding domain of the VP1 protein, but there were no variations described in GII.4-2012 variant belonging to receptor-binding site 1 (S343, T344, R345, A346, K348, N373 D374, D391, G442, and Y443) which always kept conserved except A346G occurred after 2002 [32]. N373H was a novel variant which located in the receptor-binding site, and 16 GII.4-2012 strains used Arg to replace the original Asn. Therefore, the effect of this substitution should be further verified.
Besides, five blockage epitopes (A–E) on the P2 subregion had already been well identified, and the mutations on the epitopes A (294, 296, 297, 298, 368, and 372), B (340 and 376), and E (407, 412, and 413) caused the emergence of a new GII.4 pandemic strain in response to herd immunity via positive selection [5]. Previous studies conducted by Lindesmith L. C. et al. confirmed that the variation on the epitopes A and E caused the different immunogenicity between GII.4-2002, GII.4-2006b, GII.4-2009, and the recent GII.4-2012 [33–35]. The effect of the distinguished usages of amino acid on epitopes A and E was evaluated by homology modeling. The variation caused significant changes of solvent-accessible surface areas, but did not decrease the antigenicity score. Further studies are also needed to elucidate the variations in ORF2, which can help in designing NoV vaccines.
In recent years, continuous widespread epidemic of GII.4 NoVs created an enormous social and economic burden on communities and health systems. With the development of in vitro viral reproduction system, comprehensive understanding of the virus on the basis of its whole genome became necessary to improve the accuracy of genetic classification and develop new prevention and control measures. In this study, a novel NoV GII.4-2012 strain GZ2013-L10 was confirmed based on its genome, and variations on the receptor-binding sites and blockage epitopes were also identified. The results of this study could be utilized for future genetic and evolutionary studies on NoV diversity.
References
F. Kowalzik, M. Riera-Montes, T. Verstraeten, F. Zepp, Pediatr. Infect. Dis. J. 34, 229–234 (2015)
S.M. Ahmed, A.J. Hall, A.E. Robinson, L. Verhoef, P. Premkumar, U.D. Parashar, M. Koopmans, B.A. Lopman, Lancet Infect. Dis. 14, 725–730 (2014)
S.M. Karst, C.E. Wobus, I.G. Goodfellow, K.Y. Green, H.W. Virgin, Cell Host Microbe 15, 668–680 (2014)
B.V.V. Prasad, M.E. Hardy, T. Dokland, J. Bella, M.G. Rossmann, M.K. Estes, Science 286, 287–290 (1999)
K. Debbink, L.C. Lindesmith, E.F. Donaldson, R.S. Baric, PLoS Pathog. 8, e1002921 (2012)
V. Martella, N. Decaro, E. Lorusso, A. Radogna, P. Moschidou, F. Amorisco, M.S. Lucente, C. Desario, V. Mari, G. Elia, K. Banyai, L.E. Carmichael, C. Buonavoglia, J. Virol. 83, 11391–11396 (2009)
J. Vinje, J. Clin. Microbiol. 53, 373–381 (2015)
J.S. Eden, J. Hewitt, K.L. Lim, M.F. Boni, J. Merif, G. Greening, R.M. Ratcliff, E.C. Holmes, M.M. Tanaka, W.D. Rawlinson, P.A. White, Virology 450, 106–113 (2014)
K. Bok, E.J. Abente, M. Realpe-Quintero, T. Mitra, S.V. Sosnovtsev, A.Z. Kapikian, K.Y. Green, J. Virol. 83, 11890–11901 (2009)
J.S. Noel, R.L. Fankhauser, T. Ando, S.S. Monroe, R.I. Glass, J. Infect. Dis. 179, 1334–1344 (1999)
J. van Beek, K. Ambert-Balay, N. Botteldoorn, J.S. Eden, J. Fonager, J. Hewitt, N. Iritani, A. Kroneman, H. Vennema, J. Vinje, P.A. White, M. Koopmans, NoroNet, Eurosurveillance 18, 8–9 (2013)
M.C.W. Chan, T.F. Leung, A.K. Kwok, N. Lee, P.K.S. Chan, Emerg. Infect. Dis. 20, 658–661 (2014)
J.G. Fu, J. Ai, X. Qi, J. Zhang, F.Y. Tang, Y.F. Zhu, J. Med. Virol. 86, 1226–1234 (2013)
L.P. Jia, Y. Qian, Y. Zhang, L. Deng, L.Y. Liu, R.N. Zhu, L.Q. Zhao, H. Huang, C.G. Zheng, H.J. Dong, Infect. Genet. Evol. 28, 71–77 (2014)
L. Xue, Q.P. Wu, R.M. Dong, X.X. Kou, Y.L. Li, J.M. Zhang, W.P. Guo, Foodborne Pathog. Dis. 10, 888–895 (2013)
L. Xue, Q.P. Wu, X.X. Kou, W.C. Cai, J.M. Zhang, W.P. Guo, Infect. Genet. Evol. 31, 110–117 (2015)
L. Xue, Q.P. Wu, X.X. Kou, W.C. Cai, J.M. Zhang, W.P. Guo, Virus Genes 47, 228–234 (2013)
M. Cotten, V. Petrova, M.V.T. Phan, M.A. Rabaa, S.J. Watson, S.H. Ong, P. Kellam, S. Baker, J. Virol. 88, 11056–11069 (2014)
R.A. Bull, M.M. Tanaka, P.A. White, J. Gen. Virol. 88, 3347–3359 (2007)
L. Xue, W. Cai, Q. Wu, X. Kou, J. Zhang, W. Guo, Diagn. Microbiol. Infect. Dis. (2015). doi:10.1016/j.diagmicrobio.2015.10.022
J.D. Thompson, T.J. Gibson, F. Plewniak, F. Jeanmougin, D.G. Higgins, Nucleic Acids Res. 25, 4876–4882 (1997)
K. Tamura, G. Stecher, D. Peterson, A. Filipski, S. Kumar, Mol. Biol. Evol. 30, 2725–2729 (2013)
A. Kroneman, H. Vennema, K. Deforche, H. von der Avoort, S. Penaranda, S. Oberste, J. Vinje, M. Koopmans, J. Clin. Virol. 51, 121–125 (2011)
K. Arnold, L. Bordoli, J. Kopp, T. Schwede, Bioinformatics 22, 195–201 (2006)
W. Humphrey, A. Dalke, K. Schulten, J. Mol. Graph. Model. 14, 33–38 (1996)
J. Sun, D. Wu, T.L. Xu, X.J. Wang, X.L. Xu, L. Tao, Y.X. Li, Z.W. Cao, Nucleic Acids Res. 37, W612–W616 (2009)
E. Vega, L. Barclay, N. Gregoricus, S.H. Shirley, D. Lee, J. Vinje, J. Clin. Microbiol. 52, 147–155 (2014)
A. Sabria, R.M. Pinto, A. Bosch, R. Bartolome, T. Cornejo, N. Torner, A. Martinez, M. de Simon, A. Dominguez, S. Guix, C.V.G. Stud, J. Clin. Virol. 60, 96–104 (2014)
G.M. Giammanco, S. De Grazia, V. Terio, G. Lanave, C. Catella, F. Bonura, L. Saporito, M.C. Medici, F. Tummolo, A. Calderaro, K. Banyai, G. Hansman, V. Martella, Virology 450, 355–358 (2014)
L.C. Lindesmith, E.F. Donaldson, A.D. Lobue, J.L. Cannon, D.P. Zheng, J. Vinje, R.S. Baric, PLoS Med. 5, 269–290 (2008)
R.A. Bull, P.A. White, Trends Microbiol. 19, 233–240 (2011)
M. Tan, X. Jiang, Trends Microbiol. 19, 382–388 (2011)
L.C. Lindesmith, V. Costantini, J. Swanstrom, K. Debbink, E.F. Donaldson, J. Vinje, R.S. Baric, J. Virol. 87, 2803–2813 (2013)
K. Debbink, L.C. Lindesmith, E.F. Donaldson, V. Costantini, M. Beltramello, D. Corti, J. Swanstrom, A. Lanzavecchia, J. Vinje, R.S. Baric, J. Infect. Dis. 208, 1877–1887 (2013)
L.C. Lindesmith, K. Debbink, J. Swanstrom, J. Vinje, V. Costantini, R.S. Baric, E.F. Donaldson, J. Virol. 86, 873–883 (2012)
Acknowledgments
Our study was supported by the National Natural Science Foundation of China (31271878), the China Postdoctoral Science Foundation (2014M552178), Guangdong Natural Science Foundation (2014A030313668), and the Science and Technology Planning Project of Guangdong Province (2015A020217006).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No competing financial interests exist.
Additional information
Edited by Zhen F. Fu.
Liang Xue and Weicheng Cai have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xue, L., Cai, W., Wu, Q. et al. Comparative genome analysis of a norovirus GII.4 strain GZ2013-L10 isolated from South China. Virus Genes 52, 14–21 (2016). https://doi.org/10.1007/s11262-015-1283-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-015-1283-9