Abstract
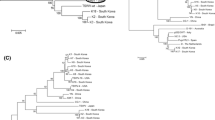
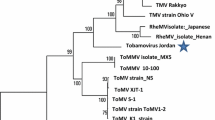
A new virus was isolated from greenhouse tomato plants showing symptoms of leaf and apex necrosis in Wielkopolska province in Poland in 2003. The observed symptoms and the virus morphology resembled viruses previously reported in Spain called Tomato torrado virus (ToTV) and that in Mexico called Tomato marchitez virus (ToMarV). The complete genome of a Polish isolate Wal’03 was determined using RT-PCR amplification using oligonucleotide primers developed against the ToTV sequences deposited in Genbank, followed by cloning, sequencing, and comparison with the sequence of the type isolate. Phylogenetic analyses, performed on the basis of fragments of polyproteins sequences, established the relationship of Polish isolate Wal’03 with Spanish ToTV and Mexican ToMarV, as well as with other viruses from Sequivirus, Sadwavirus, and Cheravirus genera, reported to be the most similar to the new tomato viruses. Wal’03 genome strands has the same organization and very high homology with the ToTV type isolate, showing only some nucleotide and deduced amino acid changes, in contrast to ToMarV, which was significantly different. The phylogenetic tree clustered aforementioned viruses to the same group, indicating that they have a common origin.



Similar content being viewed by others
References
European Plant Protection Organisation (EPPO) Reporting Service 7, 2007/128 http://archives.eppo.org/EPPOReporting/2007/Rse-0707.pdf
M. Verbeek, A.M. Dullemans, J.F.J.M. Van Den Heuvel, P.C. Maris, R.A.A. Van Den Vlugt, Arch. Virol. 152(5), 881–890 (2007). doi:10.1007/s00705-006-0917-6
M. Verbeek, A.M. Dullemans, J.F.J.M. Van Den Heuvel, P.C. Maris, R.A.A. Van Den Vlugt, Arch. Virol. 153(1), 127–134 (2007). doi:10.1007/s00705-007-1076-0
O. Le Gall, H. Sanfacon, M. Ikegami, T. Iwanami, T. Jones, A. Karasev et al., Arch. Virol. 152, 1767–1774 (2007). doi:10.1007/s00705-007-1015-0
H. Pospieszny, N. Borodynko, A. Obrepalska-Steplowska, B. Hasiów, Plant Dis. 91, 1364 (2007). doi:10.1094/PDIS-91-10-1364A
J. Sambrook, E.F. Fritsch, T. Maniatis (eds.), Molecular Cloning. A Laboratory Manual, 2nd edn. (Cold Spring Harbor Laboratory Press, New York, 1989), pp. 7.43–7.45
U.K. Laemmli, Nature 277, 680–685 (1970). doi:10.1038/227680a0
S.F. Altschul, T.L. Madden, A.A. Schäffer, J. Zhang, Z. Zhang, W. Miller et al., Nucleic Acids Res. 25, 3389–3402 (1997). doi:10.1093/nar/25.17.3389
J.D. Thompson, T.J. Gibson, F. Plewniak, F. Jeanmougin, D.G. Higgins, Nucleic Acids Res. 25(24), 4876–4882 (1997). doi:10.1093/nar/25.24.4876
K.B. Nicholas, H.B. Nicholas Jr., D.W. Deerfield II, EMBNEW. News. 4, 14 (1997)
T. Hall, BioEdit. Biological sequence alignment editor for Win95/98/NT/2 K/XP. http://www.mbio.ncsu.edu/BioEdit/bioedit.html, 2004
K. Tamura, J. Dudley, M. Nei, S. Kumar, Mol. Biol. Evol. 24(8), 1596–1599 (2007). doi:10.1093/molbev/msm092
A.R. Mushegian, Arch. Virol. 135, 437–441 (1994). doi:10.1007/BF01310028
T. Tian, L. Rubio, H.H. Yeh, B. Crawford, B.W. Falk, J. Gen. Virol. 80, 111–1117 (1999)
A.V. Karasev, Annu. Rev. Phytopathol. 38, 293–324 (2000). doi:10.1146/annurev.phyto.38.1.293
C.Y. Wu, C.F. Lo, C.J. Huang, H.T. Yu, C.H. Wang, Virology 294, 312–323 (2002). doi:10.1006/viro.2001.1344
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence of the Wal’03 isolate reported in this article has been submitted to the GenBank and is available under accession numbers: EU563948 (RNA1) and EU563947 (RNA2).
Rights and permissions
About this article
Cite this article
Budziszewska, M., Obrepalska-Steplowska, A., Wieczorek, P. et al. The nucleotide sequence of a Polish isolate of Tomato torrado virus. Virus Genes 37, 400–406 (2008). https://doi.org/10.1007/s11262-008-0284-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-008-0284-3