Abstract
The expression of antifungal genes from Trichoderma harzianum, mainly chitinases, has been used to confer plant resistance to fungal diseases. However, the biotechnological potential of glucanase genes from Trichoderma has been scarcely assessed. In this research, transgenic strawberry plants expressing the β-1,3-glucanase gene bgn13.1 from T. harzianum, under the control of the CaMV35S promoter, have been generated. After acclimatization, five out of 12 independent lines analysed showed a stunted phenotype when growing in the greenhouse. Moreover, most of the lines displayed a reduced yield due to both a reduction in the number of fruit per plant and a lower fruit size. Several transgenic lines showing higher glucanase activity in leaves than control plants were selected for pathogenicity tests. When inoculated with Colletotrichum acutatum, one of the most important strawberry pathogens, transgenic lines showed lower anthracnose symptoms in leaf and crown than control. In the three lines selected, the percentage of plants showing anthracnose symptoms in crown decreased from 61 % to a mean value of 16.5 %, in control and transgenic lines, respectively. Some transgenic lines also showed an enhanced resistance to Rosellinia necatrix, a soil-borne pathogen causing root and crown rot in strawberry. These results indicate that bgn13.1 from T. harzianum can be used to increase strawberry tolerance to crown rot diseases, although its constitutive expression affects plant growth and fruit yield. Alternative strategies such as the use of tissue specific promoters might avoid the negative effects of bgn13.1 expression in plant performance.
Similar content being viewed by others
Introduction
Strawberry (Fragaria × ananassa Duch.) is the most important soft fruit, cultivated throughout many regions of the world. A main constrain to strawberry cultivation is the low tolerance of this species to fungal diseases (Garrido et al. 2011). The number of phytopathogenic fungi that attack this crop is extensive, more than 50 genera (Mass 1998), causing severe economic losses each year. Fungal diseases can affect all parts of the strawberry plant, but those producing crown rot leading to wilting and plant dead, such as anthracnose caused by Colletotrichum spp., are among the most devastating. Control of strawberry fungal diseases is achieved mainly by chemical treatments of soil and plants. Along this line, strawberry cultivation requires an almost complete soil sterilization to get optimal fruit yield (Sjulin 2003). The most common cultural practice to get this sterilization has been the preplant soil fumigation with methyl bromide, but its use is currently forbidden in developing countries due to its harmful effect on the troposphere ozone. An approach to make this culture more environmental friendly and sustainable is the introduction of varieties resistant to fungal pathogens, and this is a major goal in most strawberry breeding programs. However, although some level of resistance has been found within the strawberry germplasm for some diseases, such as Verticillium wilt (Shaw et al. 1996) or anthracnose (Casado-Díaz et al. 2006), completely resistant genotypes have not been released.
Biotechnological tools such as genetic transformation can help to get strawberry genotypes with enhanced resistance to main fungal pathogens. Vellicce et al. (2006) obtained transgenic plants, cv. ‘Pájaro’, expressing a chitinase gene from Phaseolus vulgaris, a glucanase or a thaumatin-like protein, both from Nicotiana tabacum, or combination of these genes. Two transgenic lines expressing only the chitinase gene showed enhanced tolerance to Botrytis cinerea, but these plants were not resistant to Colletotrichum acutatum. Chitinase genes from Lycopersicon chilense and rice have also been employed to get transgenic plants with enhanced resistance to Verticillium dahliae (Chalavi et al. 2003) or Sphaerotheca humuli (Asao et al. 1997), respectively. Finally, Schestibratov and Dolgov (2005) transformed strawberry plants with the thaumatin II gene from Thaumatococcus daniellii observing some level of resistance to B. cinerea in in vitro assays.
Many attempts to increase plant protection against pathogens by the overexpression of plant chitinases or glucanases have been performed (Ceasar and Ignacimuthu 2012). However, in most cases, the increase on resistance achieved when using this strategy is effective within a narrow range of pathogens and quantitatively modest (Dana et al. 2006). Alternatively, the use of heterologous genes of well-known antipathogenic effect from fungal or bacterial origin has proven to be a more successful strategy. Trichoderma are a group of saprophytic soil fungi widely distributed in nature, which grow relatively well in different substrates without affecting animals or higher plants. Trichoderma species show an antagonistic action against phytopathogenic fungi by means of antibiotics and cell wall degrading enzymes, e.g. chitinases, glucanases, mannanases and others (Papavizas 1985; Markovich and Kononova 2003). Along this line, purified chitinases and glucanases isolated from Trichoderma have shown a very strong inhibitory action against several filamentous fungi (Lorito et al. 1993, 1994). The transgenic expression of genes encoding Trichoderma harzianum endo-chitinases, such as CHIT42 or CHIT33, increased fungal tolerance in potato (Lorito et al. 1998), apple (Bolar et al. 2001), broccoli (Mora and Earle 2001), carrot (Baranski et al. 2008) and lemon (Distefano et al. 2008). Furthermore, Dana et al. (2006) also observed an increased tolerance to salinity and heavy metals when overexpressing these genes in tobacco. Contrary to the ample information available about the expression of Trichoderma chitinases in plants, little is known about the effect of glucanases in the control of fungal diseases. The main goal of this research was to investigate the effect of the overexpression of β-1,3-glucanase gene bgn13.1 from T. harzianum on strawberry plant performance and tolerance to crown rot diseases.
Materials and methods
Plant material and Agrobacterium mediated transformation
Leaf disks from strawberry (Fragaria × ananassa, Duch., cv. ‘Camarosa’) plants micropropagated in a modified Murashige and Skoog (1962) MS medium with N30K macroelements formulation (Margara 1984) and supplemented with 2.21 µM kinetin, were used as the source of explants for genetic transformation, as previously described by Barceló et al. (1998). Briefly, leaf explants were cultured for 8 days in the shoot regeneration medium, modified MS with N30K macroelements formulation and supplemented with 8.88 µM benzyladenine (BA) and 2.46 µM indole-butyric acid (IBA), and then, inoculated with an A. tumefaciens culture diluted to a OD600 = 0.2. After 2 days of co-culture, inoculated explants were washed with sterile water and transferred to the selection medium, i.e. the regeneration medium supplemented with 50 mg l−1 kanamycin and 500 mg l−1 carbenicillin. Regenerated shoots were micropropagated and rooted under this selection pressure until acclimatization to ex vitro conditions.
The binary vector pBINB13 was used to express the β-1,3-glucanase gene bgn13.1 from T. harzianum in strawberry plants. To make this construct, the 5′ region of the cDNA from bgn13.1 gene (de la Cruz et al. 1995) was replaced by the 5′ region of the PR protein P1-p14 from tomato (Tornero et al. 1993), containing a 24 amino acids signal peptide for apoplastic localization of the mature protein. This chimaeric gene was cloned into the binary vector pBIN19 under the control of the constitutive promoter CaMV35S. The plasmid pBINB13 was introduced into A. tumefaciens, LBA4404 strain, by electroporation.
Molecular analysis of transgenic plants
The transgenic nature of the strawberry plants was confirmed by both polymerase chain reaction (PCR) amplification and Southern blot analysis. Genomic DNA was isolated from young leaves of control and putative transgenic plants growing in the greenhouse, as previously described by Manning (1991). Aliquots of these DNA extracts were used to amplify by PCR a 700 bp fragment belonging to the nptII gene, as described by Álvarez et al. (2004). For Southern blot analysis, 12 µg of DNA were digested overnight with EcoRV and HindIII, fractionated in a 0.8 % agarose gel, transferred to a nylon membrane by upward capillary transfer and crosslinked by UV irradiation. Pre-hybridization and hybridization stages were carried out at 65 °C. The filter was hybridized with a 848 bp digoxigenin labelled probe obtained by PCR amplification of the bgn13.1 gene from pBINB13 plasmid using primers β-1-3F (atcagtgctcaagacc) and β-1-3R (gcaaccttattctggc).
Phenotypical analysis of transgenic plants
After acclimatization, transgenic mother plants were propagated by runners until obtaining 8 daughter plants per line. Runners were potted individually in 12 cm pots and after 2–3 months of culture, plants were transferred to 22 cm diameter pots containing a mixture of peat moss, sand and perlite (6:3:1). These plants were grown in the greenhouse until fruiting, under natural temperature and photoperiod. Non-transformed plants conventionally propagated by runners were used as controls.
Plant surface was estimated assuming that the plant acquired an ellipse-like form by measuring long (a) and short (b) axis (area = π × a/2 × b/2). Length of petioles from fully expanded leaves was also recorded. Fruits were harvested at the stage of full ripeness, 100 % surface red, and fruit yield recorded as the total number of fruits per plant and g of fruits per plant.
Glucanase activity
β-1,3-glucanase activity was measured in young expanding leaves from control and transgenic plants growing in the greenhouse by determining the release of reducing sugars from laminarin. Frozen leaf material, 0.2 g, was ground to powder and extracted with 1 ml of 50 mM acetate buffer, pH 5.5. Extracts were centrifuged at 20,000g for 15 min and desalted using a Sephadex 50 column. The reaction mixture, containing 150 µl of 0.5 % (w/v) laminarin in extraction buffer and 50 µl desalted extract, was incubated at 37 °C for 20 h. Then, the amount of reduced sugars was estimated by the cyanoacetamide method (Gross 1982) using glucose as standard. The amount of protein in leaf extracts was measured by Bradford. Three independent extractions were performed.
Fungal infection assays
For fungal infection assays, transgenic mother plants and control plants micropropagated in N30K medium were acclimated to ex vitro conditions and transplanted to 14-cm pots containing peat moss-sand (3:1) sterilised substrate. Plants were maintained in the greenhouse for 10–12 weeks before fungal inoculation.
Susceptibility to anthracnose caused by C. acutatum was evaluated according to de los Santos and Romero (1999). C. acutatum, isolate CECT 20240, was grown on potato-dextrose agar (PDA) (Difco) supplemented with 2 g l−1 yeast extract under continuous light of 75 µmol m−2 s−1, at 25 °C for 7 days. Fungal culture plates were flooded with 5 ml of sterile distilled water and the colony surface was scraped with a scalpel. The conidial suspension obtained was filtered through sterile cheesecloth and diluted to a final concentration of 104 conidia ml−1 with sterile distilled water. Twelve plants per transgenic and control lines were sprayed with the conidial suspension until runoff. Inoculated plants were grown in a growth chamber at 25 °C under a 16 h photoperiod with 100 µmol m−2 s−1 irradiance. Disease symptoms were evaluated during 6 weeks by weekly visual observation. Disease severity index (DSI) was calculated according to the 0–5 scale described by Denoyes and Baudry (1995). In this scale, DSI values in the range 0–2.5 represent petiolar and foliar necrosis and values from 3 to 5 represent wilting due to the crown rot. A full description of this scale is as follows: 0, no lesion; 0.5, lesion just visible on the petiole or foliage; 1.0, a single developed lesion on the petiole or foliage; 1.5, two lesions; 2.0, at least two leaves with expanded lesions; 2.5, stunted plant but not wilted; 3.0, beginning of wilting; 3.5, two wilted leaves; 4.0, most leaves wilted; 4.5, all leaves wilted but slightly green; 5.0, dead plant. Percentages of plants with visible symptoms in crown, leaf or petiole after the 6 week period were also recorded.
Rosellinia necatrix assays were performed as described by Ruano-Rosa and López-Herrera (2009), using wheat seeds colonized by a single isolate of the fungus as inocula. The grains were soaked in distilled water for 24 h, autoclaved at 121 °C and 0.1 MPa for 40 min and inoculated with disks from colonies of R. necatrix, isolate CH-53, grown on PDA medium. The seeds were incubated for 15 days at 24 °C in the dark. A first experiment was performed to determine the optimal level of inocula. To this purpose, 0.25, 0.5, 1 or 1.5 g colonized wheat seeds were placed in the plant substrate, uniformly distributed and close to the stem. Plants were grown in a growth chamber at 25 °C and 100 µmol m−2 s−1 for 8 weeks. Visual symptoms were observed every 3 days, using the following scale: (1) healthy plant; (2) first symptoms of wilt in the leaves; (3) wilted plant with green leaves; (4) wilted plant with first symptoms of leaf desiccation; (5) dead plant. Areas under the disease progress curve (AUDPC) were calculated as described by Campbell and Madden (1990).
Statistical analysis
Data were analyzed by ANOVA using SPSS software, version 14 (SPSS). The Levene test for homogeneity of variances was performed prior to ANOVA, and multiple mean comparisons were done by LSD. Dunnet or Mann–Whitney U tests were used for pair comparison, in case of homogeneous or non-homogeneous variances, respectively. All tests were performed at P = 0.05.
Results
Phenotypical and molecular characterization of transgenic plants
Two independent transformation experiments with the binary plasmid pBINB13 were performed following the protocol of Barceló et al. (1998). After several months of culture in the presence of kanamycin, 23 shoots derived from independent leaf explants were obtained, yielding a 5.1 % transformation rate. Twelve lines showing the best proliferation rate in the presence of kanamycin were selected for further experiments. These shoots were micropropagated according to Barceló et al. (1998) and acclimated to ex vitro conditions. The transgenic nature of plants was confirmed by PCR amplification of a 700 bp fragment belonging to the gene nptII. All transgenic lines showed the correct amplification of the transgene, whereas no signal was observed in DNA from control plants (results not shown). After acclimatization, plants were transferred to the greenhouse for phenotypical and molecular evaluation.
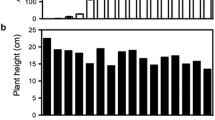
As regard to growth behaviour, some lines showed values of plant area and petiole length similar or even higher than control (Table 1). However, lines TL2, 10, 11, 15, and specially TL12 were smaller than control. The dwarf phenotypes of lines TL12 and 15 were also observed in in vitro plants. Regarding fruit yield, in general, all the lines showed a reduced yield due to a decrease in both fruit set and fruit size (Table 1). Transgenic lines could be classified in two groups. Lines TL2, 3, 9, 10, 12, 15 and 16 produced a very low number of fruits, with a mean fruit yield of 7.9 ± 6.6 g plant−1. By contrast, lines TL1, 6, 11, 21 and 23 yielded in average 68.5 ± 7.8 g plant−1, a 53 % of the fruit yield obtained in control line. Fruit weight in all the lines was also significantly lower than control, except in the case of TL21. Average fruit weight in the group of low producer transgenic lines was also lower than the one observed in medium producer lines, 2.6 ± 1.1 versus 5.4 ± 2.5 g. This behaviour in plant growth and fruit yield was maintained in following years of analysis, when daughter plants derived from runners were cultivated in the greenhouse. Three independent lines from each group (TL2, 10 and 15 from the very low producer plants and TL1, 6, and 11 from the second group) were selected for further analyses.
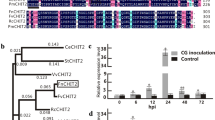
Genomic DNA extracted from leaves was subjected to Southern blot analysis using a 848 bp fragment from bgn13.1 gene as probe (Fig. 1). All selected transgenic lines showed a different pattern of hybridization bands, ranging the number of gene copies inserted between 1 in lines TL1 and 11, to 4 in the case of TL2. Glucanase activity was measured in leaf protein extracts from control and selected transgenic lines. Extracts from all lines assayed displayed significantly higher glucanase activity than control line (Fig. 2). The increments on glucanase activity were similar in all transgenic lines, about three times higher than the control, except for the line TL10 that only showed a two fold increase on activity.
Southern blot analysis of DNA from control and plants transformed with the plasmid pBINB13. DNA was digested with EcoRV and HindIII and the filter was hybridized with a digoxigenin labelled probe obtained by PCR amplification of a 848 bp sequence from pBINB13 plasmid. TL: DNA from transgenic lines. Each lane corresponds to an independent transgenic line; C: DNA from control, non-transformed plant; P: plasmid pBINB13; MW: molecular weight marker
Colletotrichum infection assays
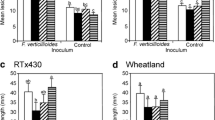
The screening for tolerance to C. acutatum of all the independent bgn13.1 transgenic lines obtained, indicated that 7 out of 12 lines displayed some level of resistance when measuring the appearance of anthracnose symptoms in crown, leaf and petioles. Based on this screening, a more detailed analysis of anthracnose resistance was performed in the most resistant selected lines, TL2, 6 and 11 (Table 2). In the three transgenic lines, the percentages of plants showing anthracnose symptoms in crown and leaves were significantly lower than in control. It is remarkable the striking reduction in the percentage of plants showing infection symptoms in the crown in the transgenic lines, being the average percentage of the three lines of 16.5 %, a value three times lower than the one observed in control. By contrast, the presence of anthracnose symptoms in petioles was only significantly reduced in the line TL11. In the three transgenic lines, DSI values were significantly lower than in the control (Table 2). Globally, the highest C. acutatum resistance was observed in the line TL11, which showed the lowest percentages of plants with anthracnose symptoms in crown, leaves and petioles, and, also the lowest DSI value. When considering the results obtained in the six transgenic lines selected, the leaf glucanase activity correlated negatively with DSI values (Pearson coefficient r = −0.771, significant at P = 0.05).
Rosellinia necatrix infection assays
Initially, a preliminary experiment was performed to determine the optimal R. necatrix inocula level for strawberry plants using a scale of symptoms from 1, healthy plant, to 5, dead plant (Fig. 3a). The highest inocula, 1 and 1.5 g colonized wheat seeds, induced 100 % plant death very rapidly, 10 days after inoculation. In the case of 0.5 g inocula, this level of damage was achieved 19–20 days after inoculation. Finally, the lowest inocula, 0.25 g, did not induce the dead of the plants, reaching a value of damage slightly higher than 4 (results not shown). According to this experiment, inoculum of 0.5 g was chosen for further experiments with selected transgenic plants. In these experiments, the level of disease resistance was estimated by the area under the disease progress curve (AUDPC). Transgenic lines TL6, TL10 and TL15 displayed higher tolerance to Rosellina necatrix than control, whereas lines TL1, TL2 and TL11 showed similar AUDPC values than control (Fig. 4). The highest tolerance to this pathogen was found in line TL6 (Fig. 3b). These plants were tested in two additional experiments obtaining a similar result, i.e. AUDPC values were significantly lower than control. In all experiments, TL6 plants showed a score of damage in the range 3–4 whereas Rosellinia infection induced the dead of control non-transgenic plants at the end of the experiment.
a Scale of Rosellinia necatrix symptoms in inoculated control plants. 1 healthy plant, 2 first symptoms of wilt in the leaves, 3 advanced wilting, plants with leaves still green, 4 wilted plant with desiccated leaves, 5 dead plant. b Aspect of control (C) and transgenic strawberry plants (line TL6) after inoculation with R. necatrix
Discussion
The micoparasite fungus T. harzianum is a potential source of genes for improving plant resistance to fungal pathogens through their heterologous expression in transgenic plants. Along this line, chitinase genes from T. harzianum have successfully been employed to increase tolerance to a broad range of pathogens in several species (Markovich and Kononova 2003). However, there is little information about the antifungal effect of Trichoderma β-1,3-glucanase genes when overexpressed in plants, despite their potential effect in vitro. As far as we know, this strategy has only been assessed in rice and pear millet plants, using in both cases the Trichoderma β-1,3-glucanase gene gluc78 (Liu et al. 2007; O’Kennedy et al. 2011). In rice, susceptibility to fungal pathogens of transgenic lines was not reported (Liu et al. 2007). In pear millet, the expression of this gene unexpectedly increased susceptibility to Sclerospora graminicola infection in most transgenic lines, although one transgenic event showed some level of resistance (O’Kennedy et al. 2011). In this research, we have expressed the gene bgn13.1 from T. harzianum CECT 2413 in strawberry plants. This gene encodes a β-1,3-glucanase which hydrolyses in vitro yeast and fungal cell walls (de la Cruz et al. 1995). The constitutive expression of bgn13.1 improved strawberry tolerance to crown rot diseases caused by C. acutatum or R. necatrix in several transgenic lines.
All transgenic lines analysed showed an increased β-1,3-glucanase activity in the leaf but no significant correlation between activity and transgene copy number was observed. Lines with 3 or 4 bgn13.1 copies showed similar or even lower activity than lines with 1–2 copies. This result could be related to gene co-suppression. Vellicce et al. (2006) observed that several transgenic strawberry lines containing more than 3 copies of a chitinase or a thaumatin-like gene did not express the transgenes. Similarly, the beneficial effect of rolC expression on increasing strawberry plant productivity and Phytophthora cactorum tolerance was observed in transgenic lines containing 1 or 2 rolC copies but not in a line with five copies of the gene (Landi et al. 2009).
Anthracnose caused by C. acutatum Simmonds, is a major disease of the cultivated strawberry in Europe, where C. acutatum is subjected to statutory quarantine requirements (Denoyes-Rothan et al. 2005). Symptoms include stolons and petioles lesions, fruit rot, flower blight, leave spots and crown rot inducing wilting and plant death. Strawberry cultivars vary in their susceptibility to the anthracnose fungus and no cultivars are known to be resistant to all Colletotrichum species (Howard et al. 1992). ‘Camarosa’, the genotype used in this research, has been classified as susceptible to C. acutatum (Casado-Díaz et al. 2006), C. fragariae and C. gloeosporioides (Mackenzie et al. 2006). A preliminary evaluation of some bgn13.1 transgenic lines showed a certain level of tolerance to this pathogen in some of the lines (Mercado et al. 2007). A more complete analysis of these transgenic plants has been performed in this research. The three selected lines assessed showed lower lesions in all tissues analyzed, i.e. crown, leaf and petiole, as well as significantly lower DSI values than control plants after inoculation with the pathogen. Casado-Díaz et al. (2006) analysed gene expression in response to C. acutatum inoculation in several strawberry cultivars differing in anthracnose susceptibility. Interestingly, the β-1,3-glucanase gene Faβgln-1 was down-regulated in fruit tissues after fungal infection in cv. ‘Camarosa’. Two additional β-1,3-glucanase genes have been isolated from strawberry plants cv. ‘Chandler’ (Shi et al. 2006), a genotype that shows similar C. acutatum susceptibility than ‘Camarosa’ (Casado-Díaz et al. 2006). The expression of both genes was highly induced after C. fragariae inoculation; however, C. acutatum treatment only induced a slight up-regulation of both genes after 48 h from inoculation (Shi et al. 2006). Although plant β-1,3-glucanase genes are generally induced after fungal infection (Balasubramanian et al. 2012), this does not seem to be the case for C. acutatum and strawberry interaction. It might be therefore possible that the increased tolerance of bgn13.1 transgenic plants to C. acutatum inoculation would just be due to an increase on total glucanase activity. However, a previous research showed that the expression of a N. tabacum glucanase gene, gln2, in strawberry did not modify anthracnose susceptibility despite the correct expression of the gene (Vellicce et al. 2006). More probably, the improved behaviour of bgn13.1 transgenic plants when challenged with C. acutatum could be due to the higher antifungal activity of fungal glucanases with respect to their plant counterparts, as generally occurs with chitinases (Shakhbazau and Kartel 2008).
White root rot caused by R. necatrix is one of the most destructive diseases of fruit trees, e.g. apple, citrus, grapevines, olive, avocado, but it also affects many other species (Sztejnberg and Madar 1980; Pliego et al. 2012). This disease can be also very destructive on strawberries planted in former sites of affected orchard trees (Mass 1998). Expression of bgn13.1 glucanase gene in strawberry increased the tolerance to this pathogen in three out of six transgenic lines analysed. The most R. necatrix tolerant line, TL6, also showed a high tolerance to C. acutatum. However, this was not the case for other lines that showed similar (TL2) or slightly lower (TL11) AUDPC values than control line after R. necatrix inoculation. The lack of correlation between C. acutatum and R. necatrix tolerance is not unexpected. Baranski et al. (2008) also observed a different behaviour of individual transgenic carrot clones expressing a T. harzianum chitinase when challenged to three different pathogens. The different pathogenicity mechanism and/or the activation of different defence pathways depending of the pathogen used might explain these results. Nevertheless, our results indicate that it is possible to get transgenic lines with a high level of tolerance to both C. acutatum and R. necatrix. Furthermore, despite the increasing interest in this pathogen as an emergent threat to many crops (Pliego et al. 2012), this is the first report on the biotechnological improvement of plant tolerance to R. necatrix through genetic transformation.
Negative side effects of constitutive bgn13.1 expression in transgenic strawberry were the reduction in plant growth and fruit set in most lines. Many others strawberry lines have been obtained using the same transformation protocol than the one used in this research, without reporting noticeable effects on plant growth (Barceló et al. 1998; Quesada et al. 2009; Yousseff et al. 2013). Thus, rather than the in vitro regeneration phase, the reduced vigour and yield of transgenic bgn13.1 plants should be ascribed to the glucanase activity interfering with cell growth and/or pollination. In fact, fruit yield of the bgn13.1 transgenic lines correlated negatively with leaf glucanase activity (Pearson coefficient r = −0.692, significant at P = 0.1). Liu et al. (2007) also observed a reduced growth in transgenic rice plants expressing the glucanase gluc78 gene from Trichoderma. Besides their role in plant defense, β-1,3-glucans also play pivotal roles in various physiological processes associated with plant growth and development, such as male gametophyte development, pollination, seed germination or stress protection (Pirselová and Matusiková 2013). Callose is also the main component on the forming cell plate (Pirselová and Matusiková 2013). Constitutive overexpression of highly active glucanases from Trichoderma might modify early cell wall deposition, interfering with cell growth. These enzymes may also release cell wall oligomers that can act as elicitors to trigger plant defense mechanisms (Hématy et al. 2009). As an alternative explanation, the reduced growth in transgenic plants may be due to an altered metabolism due to the constitutive activation of pathogen response pathways.
In conclusion, the expression of the Trichoderma glucanase gene bgn13.1 in strawberry enhances plant tolerance to crown rot diseases caused by C. acutatum and R. necatrix. However, the increased glucanase activity of transgenic lines resulted in a reduction in plant growth and/or fruit yield. Therefore, to use this gene in biotechnological breeding programs, alternative strategies should be tested to address this problem, i.e. use of tissue-specific promoters or constitutive promoters weaker than CaMV35S.
References
Álvarez R, Alonso P, Cortizo M, Celestino C, Hernández I, Toribio M, Ordás RJ (2004) Genetic transformation of selected mature cork oak (Quercus suber L.) trees. Plant Cell Rep 23:218–223
Asao H, Nishizawa Y, Arai S, Sato T, Hirai M, Yoshida K, Shinmyo A, Hibi T (1997) Enhanced resistance against a fungal pathogen Sphaerotheca humuli in transgenic strawberry expressing a rice chitinase gene. Plant Biotechnol 14:145–149
Balasubramanian V, Vashisht D, Cletus J, Sakthivel N (2012) Plant β-1,3-glucanases: their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol Lett 34:1983–1990
Baranski R, Klocke E, Nothnagel T (2008) Chitinase CHIT36 from Trichoderma harzianum enhances resistance of transgenic carrot to fungal pathogens. J Phytopathol 156:513–521
Barceló M, El Mansouri I, Mercado JA, Quesada MA, Pliego-Alfaro F (1998) Regeneration and transformation via Agrobacterium tumefaciens of the strawberry cultivar Chandler. Plant Cell Tissue Organ Cult 54:29–36
Bolar JP, Norelli JL, Harman GE, Brown SK, Aldwinkle HS (2001) Synergistic activity of endochitinase and exochitinase from Trichoderma atroviride (T. harzianum) against the pathogenic fungus (Venturia inaequalis) in transgenic apple plants. Transgenic Res 10:533–543
Campbell CL, Madden LV (1990) Temporal analysis of epidemics. I: descriptions and comparisons of disease progress curve. In: Campbell CL, Madden LV (eds) Introduction to plant disease epidemiology. Wiley, New York, pp 161–202
Casado-Díaz A, Encinas-Villarejo S, Santos BDL, Schilirò E, Yubero-Serrano E-M, Amil-Ruíz F, Pocovi MI, Pliego-Alfaro F, Dorado G, Rey M, Romero F, Muñoz-Blanco J, Caballero JL (2006) Analysis of strawberry genes differentially expressed in response to Colletotrichum infection. Physiol Plant 128:633–650
Ceasar A, Ignacimuthu S (2012) Genetic engineering of crop plants for fungal resistance: role of antifungal genes. Biotechnol Lett 34:995–1002
Chalavi V, Tabaeizadeh Z, Thibodeau P (2003) Enhanced resistance to Verticillium dahliae in transgenic strawberry plants expressing a Lycopersicon chilense chitinase gene. J Am Soc Hortic Sci 128:747–753
Dana MM, Pintor-Toro JA, Cubero B (2006) Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol 142:722–730
de la Cruz J, Pintor-Toro JA, Benítez T, Llobell A, Romero LC (1995) A novel endo-β-1,3-glucanase, BGN13.1, involved in the mycoparasitism of Trichoderma harzianum. J Bacteriol 177:6937–6945
de los Santos B, Romero-Muñoz F (1999) Occurrence of Colletotrichum acutatum, causal organism of strawberry anthracnose in Southwestern Spain. Plant Dis 83:301
Denoyes B, Baudry A (1995) Species identification and pathogenicity study of French Colletotrichum strains isolated from strawberry using morphological and cultural characteristics. Phytopathology 85:53–57
Denoyes-Rothan B, Guérin G, Lerceteau-Köhler E, Risser G (2005) Inheritance of resistance to Colletotrichum acutatum in Fragaria × ananassa. Phytopathology 95:405–412
Distefano G, La Malfa S, Vitale A, Lorito M, Deng Z, Gentile A (2008) Defence-related gene expression in transgenic lemon plants producing an antimicrobial Trichoderma harzianum endochitinase during fungal infection. Transgenic Res 17:873–879
Garrido C, Carbú M, Fernández-Acero FJ, González-Rodríguez VE, Cantoral JM (2011) New insights in the study of strawberry fungal pathogens. In: Husaini AM, Mercado JA (eds) Genomics, transgenics, molecular breeding and biotechnology of strawberry, Glob Sci Book, pp 24–39
Gross KC (1982) A rapid and sensitive spectrophotometric method for assaying polygalacturonase using 2-cyanoacetamide. HortScience 17:933–934
Hématy K, Cherk C, Somerville S (2009) Host-pathogen warfare at the plant cell wall. Curr Opin Plant Biol 12:406–413
Howard CM, Maas JL, Chandler CK, Albregts EE (1992) Anthracnose of strawberry caused by the Colletotrichum complex in Florida. Plant Dis 76:976–981
Landi L, Capocasa F, Costantini E, Mezzetti B (2009) ROLC strawberry plant adaptability, productivity, and tolerance to soil-borne disease and mycorrhizal interactions. Transgenic Res 18:933–942
Liu M, Zhu J, Sun Z-X, Xu T (2007) Possible suppression of exogenous β-1,3-glucanase gene gluc78 on rice transformation and growth. Plant Sci 172:888–896
Lorito M, Harman GE, Hayes CK, Broadway RM, Tronsmo A, Woo SL, Di Pietro A (1993) Chitinolytic enzymes produced by Trichoderma harzianum: antifungal activity of purified endochitinase and chitobiosidase. Phytopathology 83:302–307
Lorito M, Peterbauer C, Hayes CK, Harman GE (1994) Synergistic interaction between fungal cell wall-degrading enzymes and different antifungal compounds enhances inhibition of spore germination. Microbiology 140:623–629
Lorito M, Woo SL, Fernández IG, Colucci G, Harman GE, Pintor-Toro JA, Filipone E, Muccifora S, Lawrence CB, Zoina A, Tuzun S, Scala F (1998) Genes from mycoparasitic fungi as source for improving plant resistance to fungal pathogens. Proc Natl Acad Sci USA 95:7860–7865
Mackenzie SJ, Legard DE, Timmer LW, Chandler CK, Peres NA (2006) Resistance of strawberry cultivars to crown rot caused by Colletotrichum gloeosporioides isolates from Florida is nonspecific. Plant Dis 90:1091–1097
Manning K (1991) Isolation of nucleic acids from plants by differential solvent precipitation. Anal Biochem 195:45–50
Margara J (1984) Bases de la multiplication végétative. INRA, Versailles
Markovich NA, Kononova GL (2003) Lytic enzymes of Trichoderma and their role in plant defense from fungal diseases: a review. Appl Biochem Micro 39:341–351
Mass JL (1998) Compendium of strawberry diseases, 2nd edn. American Phytophatological Society Press, St Paul
Mercado JA, Martín-Pizarro C, Pascual L, Quesada MA, Pliego-Alfaro F, de los Santos B, Romero F, Gálvez J, Rey M, de la Viña G, Llobell A, Yubero-Serrano E-M, Muñoz-Blanco J, Caballero JL (2007) Evaluation of tolerance to Colletotrichum acutatum in strawberry plants transformed with Trichoderma-derived genes. Acta Hortic 738:383–388
Mora AA, Earle ED (2001) Resistance to Alternaria brassicicola in transgenic broccoli expressing Trichoderma harzianum endochitinase gene. Mol Breed 8:1–9
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
O’Kennedy MM, Crampton BG, Lorito M, Chakauya E, Breese WA, Burger JT, Botha FC (2011) Expression of a β-1,3-glucanase from a biocontrol fungus in transgenic pearl millet. S Afr J Bot 77:335–345
Papavizas GC (1985) Trichoderma and Gliocladium: biology and potential for biological control. Ann Rev Phytopathol 23:23–54
Pirselová B, Matusiková I (2013) Callose: the plant cell wall polysaccharide with multiple biological functions. Acta Physiol Plant 35:635–644
Pliego C, López-Herrera C, Ramos C, Cazorla FM (2012) Developing tools to unravel the biological secrets of Rosellinia necatrix, an emergent threat to woody crops. Mol Plant Pathol 13:226–239
Quesada MA, Blanco-Portales R, Posé S, García-Gago JA, Jiménez-Bermúdez S, Muñoz-Serrano A, Caballero JL, Pliego-Alfaro F, Mercado JA, Muñoz-Blanco J (2009) Antisense down-regulation of the FaPG1 gene reveals an unexpected central role for polygalacturonase in strawberry fruit softening. Plant Physiol 150:1022–1032
Ruano-Rosa D, López-Herrera CJ (2009) Evaluation of Trichoderma spp. as biocontrol agents against avocado white root rot. Biol Control 51:66–71
Schestibratov KA, Dolgov SV (2005) Transgenic strawberry plants expressing a thaumatin II gene demonstrate enhanced resistance to Botrytis cinerea. Sci Hortic 106:177–189
Shakhbazau AV, Kartel NA (2008) Chitinases in bioengineering research. Russ J Genet 44:881–889
Shaw DV, Gubler WD, Larson KD, Hansen J (1996) Genetic variation for field resistance to Verticillium dahliae evaluated using genotypes and segregating progenies of California strawberries. J Am Soc Hortic Sci 121:625–628
Shi Y, Zhang Y, Shih DS (2006) Cloning and expression analysis of two β-1,3-glucanase genes from strawberry. J Plant Physiol 163:956–967
Sjulin TM (2003) The North American small fruit industry 1903–2003. II. Contributions of public and private research in the past 25 years and a view to the future. HortScience 38:960–967
Sztejnberg A, Madar Z (1980) Host range of Dematophora necatrix, the cause of white root rot disease in fruit trees. Plant Dis 64:662–664
Tornero P, Rodrigo I, Conejero V, Vera P (1993) Nucleotide sequence of a cDNA encoding a pathogenesis-related protein, Pi-p14, from tomato (Lycopersicon esculentum). Plant Physiol 102:325
Vellicce GR, Ricci JCD, Hernández L, Castagnaro AP (2006) Enhanced resistance to Botrytis cinerea mediated by the transgenic expression of the chitinase gene ch5B in strawberry. Trans Res 15:57–68
Yousseff SM, Amaya I, López-Aranda JM, Sesmero R, Valpuesta V, Casadoro G, Blanco-Portales R, Pliego-Alfaro F, Quesada MA, Mercado JA (2013) Effect of simultaneous down-regulation of pectate lyase and endo-β-1,4-glucanase genes on strawberry fruit softening. Mol Breed 31:313–322
Acknowledgments
This research was supported by the Comisión Interministerial de Ciencia y Tecnología in Spain and FEDER EU Funds (Grant No. AGL2011-30354-C02-01).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mercado, J.A., Barceló, M., Pliego, C. et al. Expression of the β-1,3-glucanase gene bgn13.1 from Trichoderma harzianum in strawberry increases tolerance to crown rot diseases but interferes with plant growth. Transgenic Res 24, 979–989 (2015). https://doi.org/10.1007/s11248-015-9895-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-015-9895-3