Abstract
Chimaeras, or ratfishes, are the only extant group of holocephalan fishes and are the sole host group of gyrocotylidean cestodes, which represent a sister group of the true tapeworms (Eucestoda). These unique, non-segmented cestodes have been known since the 1850s and multiple species and genera have been erected despite a general agreement that the delineation of species on the basis of morphology is effectively impossible. Thus, in the absence of molecular studies, the validity of gyrocotylid taxa and their specific host associations has remained highly speculative. Here we report the presence of Gyrocotyle spp. from rarely-caught deep-sea chimaeras collected in the North-East Atlantic, and describe two new species: G. haffii n. sp. from the bent-nose chimaera, Harriota raleighana Goode & Bean, and G. discoveryi n. sp. from the large-eyed rabbit fish, Hydrolagus mirabilis (Collett). Nuclear ribosomal sequence data were generated for individual parasites taken from different host species collected on different dates and from different localities and were combined with previously published sequences. Phylogenetic analyses supported the recognition of independent lineages and clusters, indicative of species, but were indecisive in recovering the root of the tree in analyses that included non-gyrocotylid outgroup taxa. The molecular data reveal variation not reflected in morphology and point to a complex picture of genetic divergence shaped by both isolation and migration in the deep-sea environment.
Similar content being viewed by others
Introduction
Holocephalans are deep-sea, cartilaginous fish of the chondrichthyan subclass Holocephali and its only order, the Chimaeriformes, commonly known as rat fishes or ghost sharks. Although highly successful in the Palaeozoic, resulting in a rich palaeontological fauna, the group is now represented by only 39 species in five genera (Inoue et al., 2010). Molecular studies confirm that the group is the sister taxon of the Elasmobranchii and three families are recognised, the Callorhinchidae, Chimaeridae and Rhinochimaeridae, where the former is the sister group to the latter two (Inoue et al., 2010). According to a relaxed molecular clock method employed by Inoue et al. (2010), the Holocephali arose in the Silurian Period (c.410–447 Ma), the Callorhinchidae diverged from its sister group in the Jurassic Period (c.161–190 Ma) and the other families diverged in the mid-Cretaceous Period (c.98–146 Ma). Licht et al. (2012) expanded the representation of holocephalans and reported similar results, with the group diverging between the late Silurian and the early Devonian. This ancient and distinctive host group harbours similarly distinctive parasites, and in this study, we report on those of the cestode order Gyrocotylidea.
Gyrocotylideans are non-segmented tapeworms known together with the Amphilinidea as cestodarians and are the putative sister group of the true tapeworms, or eucestodes (Waeschenbach et al., 2012). Within the order, species of the only accepted genus Gyrocotyle Diesing, 1850 are common, well-reported parasites of holocephalans. Their systematics have been reviewed previously (e.g. Colin et al., 1986; Bandoni & Brooks, 1987; Williams et al., 1987; Gibson, 1994) and the morphological characters for distinguishing species have been thoroughly discussed and, according to Williams et al. (1987), been found wanting. Moreover, in addition to the lack of reliable morphological characters for species identification, the method of preservation has been shown to have a significant effect on their morphology, making it difficult to provide reliable species identifications post-preservation (Colin et al., 1986). In the absence of such characters, it appears that many specimens have been identified historically on the basis of their host species. Meanwhile, sequence data are available for only three putative species.
In this paper we use partial large nuclear ribosomal subunit (lsrDNA; domains D1–D3) and complete small nuclear ribosomal subunit (ssrDNA) sequences to reconstruct a phylogenetic network of Gyrocotyle spp. from five chimaeras: Chimaera monstrosa L., Hydrolagus mirabilis (Collett), Hydrolagus colliei (Lay & Bennett) (Chimaeridae), Harriotta raleighana Goode & Bean (Rhinochimaeridae) and Callorhinchus milii Bory de Saint-Vincent (Callorhinchidae). Based on these data, we recognise and name two new species of Gyrocotyle from the bent-nose chimaera, Ha. raleighana, and the large-eyed rabbit fish, Hy. mirabilis. These deep-sea host species are not commonly seen and, although probably not rare at depths greater than 1,000 metres, are difficult to capture, requiring specialised equipment and considerable effort.
Materials and methods
Specimen collection, preservation and morphological study
Chimaeras were collected by RAB using a semi-balloon otter trawl during three research cruises in the North-East Atlantic aboard the National Environmental Research Council research vessel RRS Discovery (April 2001 and September/October 2002). The fishes were immediately dissected and worms extracted and fixed briefly in Berland′s fluid and preserved in 80% ethanol for morphological examination, and in 100% ethanol for molecular analysis. In some cases, worms were cut into separate parts before fixation. Whole-mounts were stained with Mayer′s paracarmine, cleared in beechwood creosote and mounted in Canada balsam. Measurements were made through a drawing tube on an Olympus BH-2 microscope using a Digicad Plus digitising tablet and Carl Zeiss KS100 software adapted by Imaging Associates, and are quoted in micrometres. Where two-dimensions are given length precedes width. Type- and voucher material has been submitted to the Natural History Museum, London, UK (NHMUK). Additional specimens for molecular analysis were obtained from waters off Norway (Tromsø, Finnmark and Bergen) and Tasmania, Australia, and published sequences of three Gyrocotyle species were included in the analyses. A list of taxa including collection information and sequence accession numbers is given in Table 1.
Molecular analysis
Ethanol was removed from tissue samples by soaking in tris-EDTA buffer overnight or by evaporation at room temperature. Total genomic DNA was extracted using the DNeasy blood and tissue kit (Qiagen). Partial lsrDNA (domains D1-D3; c.1,400 bp) was amplified using LSU5 or ZX-1 + 1200R or 1500R primers; in the case of Gyrocotyle confusa van der Land & Dienske, 1968, only a short fragment of 512 bp could be sequenced from a fragment amplified using primers 900F + 1500R. Complete ssrDNA (c.2,000 bp) was amplified using WormA and WormB primers for a subset of the taxa (see Table 1). PCRs were carried out in 25 µl reaction volumes using puRe Taq Ready-to-go PCR beads (Amersham Biosciences, Little Chalfont, UK) and 1 µl of a 10 µM solution of each primer. Cycling conditions included an initial denaturation for 5 min at 95 °C, followed by 40 cycles of 30 s at 95 °C denaturation, 30 s at 55 °C (lsrDNA) or 54°C (ssrDNA) and 2 min at 72 °C, followed by a final hold of 7 min at 72 °C. Amplicons were purified using a QIAquick Gel Extraction Kit or a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). Sequencing of both strands was carried out on an Applied Biosystems 3730 DNA Analyser, using Big Dye version 1.1. PCR and internal sequencing primers for lsrDNA are given in Littlewood et al. (2000), except for ZX-1 which was modified from van der Auwera et al. (1994) as shown in bold: ACC CGC TGA ATT TAA GCA TAT. Primers for ssrDNA are given in Littlewood & Olson (2001). Contigs were assembled using Sequencher 4.5 (GeneCodes Corporation, Ann Arbor, USA) and manually checked for ambiguous and incorrect base calls. Sequence identity was verified using the Basic Local Alignment Search Tool (BLAST) (www.ncbi.nih.gov/BLAST/).
Phylogenetic analysis
Gene-specific alignments were made for all available gyrocotylidean sequences along with multiple representatives of either caryophyllidean or spathebothriidean species (see Olson et al., 2008) used as outgroups. In addition, a gyrocotylidean-only sequence alignment was made. Sequences were aligned with MAFFT version 7.149b (Katoh, 2008) using 1,000 cycles of iterative refinement and the genafpair algorithm. Alignment masks for ambiguously aligned positions were generated using GBLOCKS (Castresana, 2000; Talavera & Castresana, 2007) using less stringent settings, and were further refined by eye in Mesquite version 3.5 (Maddison & Maddison, 2018). Alignments with indicated exclusion sets are available from the NHM Data Portal at https://doi.org/10.5519/0003327. MrModeltest2 (Nylander, 2004) was used to select a model of nucleotide substitution using Akaikeʼs information criterion. Data were partitioned into three character sets: (i) partial lsrDNA; (ii) complete ssrDNA; and (iii) partial lsrDNA + complete ssrDNA. Phylogenetic trees were constructed using Bayesian inference with MrBayes, version 3.2 (Ronquist & Huelsenbeck, 2003). Likelihood settings were set to nst = 6, rates = invgamma, ngammacat = 4 (equivalent to the GTR+I+G model of evolution). In the combined analysis, parameters were estimated separately for each gene. Four chains (temp = 0.2) were run for 15,000,000 generations and sampled every 1,000th generation; 10,000,000 generations were discarded as ‘burn-inʼ. The ‘burn-inʼ period was determined as the point when the average standard deviation of split frequency values were < 0.01.
To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN, 2012), details of all new taxa have been submitted to ZooBank. For each new taxon, the Life Science Identifier (LSID) is reported in the taxonomic summary.
Results
Molecular analyses
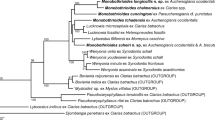
Bayesian inference analysis of the combined ssr/lsrDNA data is shown in Fig. 1, and the results of analyses of the individual gene partitions are given in Supplementary Figures S1 and S2. Nodes supported by < 0.95 posterior probabilities were collapsed. Table 2 gives the corrected (GTR+I+G) pairwise distances estimated for each gene. Analyses including either caryophyllidean or spathebothriidean outgroup taxa failed to robustly resolve relationships among the gyrocotylidean samples, as a consequence of the need to exclude large numbers of sites that lacked clear positional homology between ingroup and outgroup sequences. For example, the lsrDNA alignment including caryophyllidean outgroup taxa required 60% of the sites to be excluded whereas an alignment including only gyrocotylidean sequences required only 25% and thus included a greater number of informative characters among the ingroup sequences. We therefore chose to maximise the number of informative sites by aligning the gyrocotylidean sequences to themselves and consequently present our results as un-rooted networks.
Unrooted, consensus network of Gyrocotyle species based on combined complete ssr + partial lsrDNA. Nodes supported by < 0.95 posterior probabilities have been collapsed. Boxed inset shows the topology including the full branch subtending G. nybelini. Sample labels are given in Table 1
All data partitions showed that the most divergent taxon by an order of magnitude was G. nybelini (Fuhrmann, 1931) Bandoni & Brooks, 1987 from C. monstrosa collected off Norway (Table 2); inset boxes in Fig. 1 and Supplementary Figures S1 and S2 were required to depict its full branch length relative to the other taxa. This was followed by Gyrocotyle sp. from C. milii off Australia. Among the samples collected from the North-East Atlantic, those from Hy. mirabilis formed a tight cluster with good separation from the other branches of the network and are described below as Gyrocotyle discoveryi n. sp. A specimen from Ha. raleighana similarly formed a distinct lineage in the network and has been described below as Gyrocotyle haffii n. sp. This lineage was connected in an unresolved node with G. confusa and G. nybelini, also from C. monstrosa off Norway. Samples identified as G. urna (Grube & Wagener in Wagener, 1852) Wagener, 1858 from C. monstrosa showed considerable divergence, both among Norwegian fjords and between these and the North Atlantic, whereas the sample identified as G. rugosa Diesing, 1850 from Hy. colliei from the Gulf of Alaska was closer to the G. urna samples from Norway than they are to the G. urna sample from the North-East Atlantic. The possibility that G. urna/G. rugosa represents a single, variable species is discussed below.
Class Cestoda
Order Gyrocotylidea Poche, 1926
Family Gyrocotylidae Benham, 1901
Genus Gyrocotyle Diesing, 1850
Gyrocotyle haffii n. sp.
Type-host: Harriotta raleighana Goode & Bean (Chimaeriformes: Rhinochimaeridae), bent-nosed chimaera.
Type-locality: Goban Spur (49°46′N, 12°21′W, depth 1,631–1,653 m, 22-23.iv.2001; RRS Discovery Cruise 252, No. 13951/14), North-East Atlantic.
Type-material: Holotype (NHMUK.2019.11.21.1), paratype (NHMUK.2019.11.21.2).
Site in host: Spiral intestine.
Representative DNA sequences: MN655880 (ssrDNA); MN657006 (lsrDNA, domains D1-D3).
ZooBank registration: The Life Science Identifier (LSID) for Gyrocotyle haffii n. sp. is urn:lsid:zoobank.org:act:7717F9D6-4C9D-4C59-8D0A-4C5E306B4671.
Etymology: The species is named in honour of our late colleague and friend Professor Harford ‘Haffi′ Williams in recognition of his contribution to the understanding of the Gyrocotylidea.
Description
[Based on a single intact, immature whole worm and second immature worm from which the central portion had been excised for molecular analysis; Figs. 2, 3]. With characters of the order. Body elongate with minute annular ridges; no large lateral flap. Length 23,504; greatest width near anterior extremity, 3,131. Rosette relatively small with few crenulations, 1,943 long. Anterior sucker large, oval, 1,750 × 1,223. Reproductive system immature; anlagen commences 2,852 from anterior extremity, 8,191 long; consisting of a long, narrow patch of stained tissue reaching, and a branched section passing, towards lateral margin of worm; apparently opening at c.268 from anterior extremity. Only other evidence of reproductive organs is putative vitelline glands scattered around posterior extremity of anlagen.
Images and drawings of the new Gyrocotyle species. 2, Photomicrograph of Gyrocotyle haffii n. sp. holotype (NB: the specimen is immature) ex Harriotta raleighana, Goban Spur (13951/14); 3, Line-drawing of Gyrocotyle haffii n. sp., holotype; 4, Photomicrograph of Gyrocotyle discoveryi n. sp. holotype ex Hydrolagus mirabilis (Goban Spur; 15063/103a); 5. Line-drawing of Gyrocotyle discoveryi n. sp., holotype; 6, Gyrocotyle discoveryi n. sp. paratype ex Hydrolagus mirabilis (Goban Spur; 15066/124a). Scale-bars: 10 mm
Diagnosis
Gyrocotyle haffii n. sp. can be diagnosed from other congeners on the basis of unique nucleotide characters in our rDNA alignments (listed as alignment position-nucleotide): ssrDNA: 218-T, 723-A, 746-C, 747-A, 748-G, 1,158-T, 1,654-G, 1,673-C, 2,115-C; lsrDNA: 612-T, 837-A, 875-T, 1,306-A, 1,395-C, 1,402-A, 1,501-G.
Remarks
As far as we are aware there is only one previous report of a gyrocotylidean from Ha. raleighana, the bent-nose chimaera. Parukhin (1966) reported “Gyrocotyloides nybelini Fuhrmann, 1931” in this host from the South Atlantic Ocean. Parukhin (1968) repeated this report saying (in translation) “Found in Callorhynchus capensis. Two adult parasites were found in two fish. In addition, six larvae were found in one of them. In addition to C. capensis, specimens were found in two Hariota [sic] raleighana. In both cases there were two specimens. Previously, this species was observed in the Atlantic in Chimaera monstrosa”. In addition, it seems likely that the records of ‘cestode adults′ from Ha. raleighana, Hy. mirabilis and C. monstrosa from the Rockall Trough off NW Scotland by Mauchline & Gordon (1984) refer to Gyrocotyle spp.
There is no reliable morphological character to differentiate this species or indeed any of the gyrocotylidean species circumscribed by molecular means. Therefore, the species is diagnosed by its relatively marked sequence divergence from those of recognised species.
Gyrocotyle discoveryi n. sp.
Type-host: Hydrolagus mirabilis (Collett) (Chimaeriformes: Chimaeridae), large-eyed rabbitfish.
Type-locality: Goban Spur, North-East Atlantic.
Other localities: Porcupine Seabight (51°09′N, 11°55′W, depth 1,200 m, 30.xi.2002, RRS Discovery Cruise, No. 15048-14, 15); Goban Spur (49°49′N, 11°44′W, depth 1,175–1,250 m, 27.iv.2001, RRS Discovery Cruise 252, No. 13963/17, 20, 24, 72; 49°41′N, 11°53′W, depth 1,053–1,077 m, 23.iv.2001, RRS Discovery Cruise 252, No. 13962/4; 49°47′N, 11°58′W, depth 1,240–1,360 m, 19.x.2002, RRS Discovery Cruise D266, No. 15066-124, 125; 51°09′N, 11°55′W, depth 1200 m, 30.ix.2002, RRS Discovery Cruise D266, No. 15063-103), North-East Atlantic.
Type-material: Holotype (NHMUK 2019.11.21.3), paratypes (NHMUK.2019.11.21.4-13 from Goban Spur; NHMUK.2019.11.21.14-19 from Porcupine Sea Bight).).
Site in host: Spiral intestine.
Representative DNA sequences: MN655879 and MN655881 (ssrDNA); MN657003-MN657005, MN657007- MN657009, MN657011 (lsrDNA, domains D1-D3).
ZooBank registration: The Life Science Identifier (LSID) for Gyrocotyle discoveryi n. sp. is urn:lsid:zoobank.org:act:7B028A0B-B8EB-495E-A9F1-F29DDB60B89A.
Etymology: The species is named after the RRS Discovery, the NERC research vessel on which the specimens were collected.
Description
[Based on 17 specimens; Figs. 4–6.] With characters of the order. Body relatively squat, with deeply crenulated margins, 8,634–17,586 × 4,996–8,439 (12,071 × 6,676), width 36–98 (60)% of length. Anterior sucker distinct, 1,149–1,621 (1,347) long, 596–1,022 (771) wide. Uterus large, in central part of body, 1,546–2,996 (2,232) from anterior extremity, 2,453–6,226 (4,533) long, 35–43 (38)% of body length. Rosette distinct, fairly complex, 1,797–3,092 (2,575) long, junction with soma not clear. Eggs tanned, operculate, 85–97 × 39–56 (89 × 49).
Diagnosis
Gyrocotyle discoveryi n. sp. can be diagnosed from other congeners on the basis of unique nucleotide characters in our rDNA alignments (listed as alignment position-nucleotide): ssrDNA: 176-G, 782-G, 862-G, 973-C; lsrDNA: 573-A, 800-T, 1,245-C, 1,246-C, 1,247-G, 1,360-T, 1,369-T, 1,375-C, 1,379-G, 1,382-G, 1,391-A, 1,449-T, 1,468-T, 1,477-T.
Remarks
Mauchline & Gordon (1984) reported a “cestode” in Hy. mirabilis from the Rockall Trough off NW Scotland, which is, as far as we are aware, the only possible record of a Gyrocotyle from this host. Two species of Gyrocotyle, G. major van der Land & Templeman, 1968 and G. abyssicola van der Land & Templeman, 1968, have been reported from its congener, the small-eyed rabbit fish Hydrolagus affinis (de Brito Capello) on the edges of the continental shelf off the eastern coast of Newfoundland (van der Land & Templeman, 1968). These two species are illustrated as much more elongate than our specimens, with less complex lateral wrinkling and small rosettes. The worms were recovered from frozen hosts, so the gross morphology may well not be of significance in differentiating these species. Subsequently, these two taxa have been reported from the same host species off south-western Greenland by Karlsbakk et al. (2002) and, puzzlingly, from a rhinochimaerine species, the straight-nosed rabbit fish Rhinochimaera atlantica Holt & Byrne, off the Scotian Shelf by Hogans & Hurlbut (1984). In the North-East Atlantic, R. atlantica has not been found harbouring Gyrocotyle, but it does harbour the strobilate tapeworm Chimaerocestos prudhoei Williams & Bray, 1984 and a congeneric host, the Pacific spookfish R. pacifica (Mitsukuri, 1895) also harbours a species of Chimaerocestos Williams & Bray, 1984 (see Caira et al., 1999, 2014). Other records of Gyrocotyle spp. from Hydrolagus spp. are from the Pacific Ocean (see Bandoni & Brooks, 1987).
There are no reliable morphological characters to differentiate this species or indeed any of the gyrocotylidean species circumscribed by molecular means. Therefore, the species is diagnosed by its relatively marked sequence divergence from those of recognised species.
Discussion
Colin et al. (1986) made a careful study, based on 1,361 specimens, of the morphological characters used for distinguishing species of Gyrocotyle and concluded that, due to the great contractibility of the worms, their reactions to different fixation techniques and the state of the worms at fixation (e.g. alive, dead, from frozen hosts), some characters were of limited or no value, i.e. total length and breadth, the degree of lateral crenulation, the complexity of the rosette, the distribution of body spines and the morphology of the eggs. In effect, they came to the conclusion that Gyrocotyle spp. could not be reliably identified using morphological characters. Indeed, these authors considered Gyrocotyle confusa and Gyrocotyloides nybelini as synonyms of G. urna, and the genus Gyrocotyloides Fuhrmann, 1930 as synonymous with Gyrocotyle. When an unidentified “chimaera cestodarian” was reported in the Caribbean chimaera Chimaera cubana Howell Rivero by Bunkley-Williams & Williams (2004), they reckoned that “most authors agree that only one morphologically highly variable species of cestodarian is found in chimaeras, but some confusion exists about calling it Gyrocotyle rugosa Diesing, 1850 or G. urna (Grube & Wagener, 1852)”.
Despite the difficulties in identifying Gyrocotyle spp. on the basis of morphology and the controversies in the literature relative to the specific and generic status of various morphological forms (e.g. Colin et al., 1986; Bandoni & Brooks, 1987; Williams et al., 1987), there have been few investigations utilising molecular data. Simmons et al. (1972) utilised DNA hybridisation to confirm the distinctness of four species of Gyrocotyle from the Pacific Ocean. Bristow & Berland (1988), Berland et al. (1990) and Bristow (1992), using electrophoresis, fatty acid chemistry and biological characteristics, retained three species as distinct, but did not recognise the genus Gyrocotyloides. Olson & Caira (1999) generated an ssrDNA sequence of Gyrocotyle rugosa (Grg, Table 1), Olson et al. (2001) added partial lsrDNA data for this species and generated ssrDNA and lsrDNA data for G. urna (Gyro, Table 1), and Olson et al. (2008) generated ssr/lsrDNA sequences for Gyrocotyle sp. (Gyc, Table 1). In 2007, Waeschenbach et al. completed the lsrDNA sequence of G. urna (i.e. Gyro) and in 2012 complemented this with large fragments of mitochondrial genome data (Waeschenbach et al., 2012).
Each of the latter studies were aimed at resolving higher-level interrelationships of eucestodes and did not attempt to address the interrelationships or validity of named species and genera within the order. In this paper we make a first attempt at this, using ribosomal sequences from a variety of gyrocotylids, including some identified by other workers. The inability to root the resulting trees negated the ability to define clades, but the results still provide a picture of the relative genetic distances between samples and how they are interconnected within the network. The species G. nybelini, G. confusa and G. haffii n. sp. are part of an unresolved trichotomy, but are separated by long branches from the other samples and from each other. This indicates that G. haffii n. sp. is not conspecific with G. nybelini, suggesting in turn that Parukhin (1966, 1968) may have been incorrect in reporting G. nybelini from the host Ha. raleighana.
Gyrocotyle sp. from Callorhynchus milii off Hobart, Australia, forms another long branch in the network and, on this basis, is likely to represent an undescribed species. The hosts of this lineage of Gyrocotyle are unusual chimaeras known commonly as ghost sharks or elephant fish and are restricted to the temperate coasts of Australia and New Zealand. Not deep-sea dwelling, they constitute part of the fisheries in both countries and are commonly taken, suggesting that the collection and study of their gyrocotylid parasites should make the circumscription of this putatively novel species easier than for species whose hosts are rarely obtained.
Gyrocotyle urna, with six samples from C. monstrosa clustering with ‘G. rugosa’ from Hydrolagus colliei off Alaska, may well represent a complex of similar species, or is a single widespread species in northern waters with contacts via the deep Arctic Ocean. There is also distinct divergence between G. urna specimens from different Norwegian fjords. These may be as deep as 1,300 m and, like most fjords, are deeper than the adjacent sea and generally have a sill at their mouth formed by the glacier′s terminal moraine. This topology may explain the apparent isolation of Gyrocotyle populations from different fjords as indicated by their genetics.
Gyrocotyle discoveryi n. sp., represented by seven samples from Hy. mirabilis in the North-East Atlantic, is almost genetically homogeneous. The regions of the two sites of collection are adjacent, with the Goban Spur forming the relatively shallow bank at the southern margin of the Porcupine Seabight. The samples formed a tight cluster that most likely represents a clade specific to the large-eyed rabbitfish.
As far as we are aware, the only gyrocotylids previously reported from the North-East Atlantic are the three species known from C. monstrosa, the commonly found holocephalan in the region. These are G. urna, the most commonly reported, and two rarer forms, G. confusa and G. nybelini. As stated previously, the latter species has been housed by various authors in the genus Gyrocotyloides, but this has been more commonly accepted as a synonym of Gyrocotyle (see Gibson, 1994). In contrast, our data lend some support to the recognition of Gyrocotyloides as a distinct genus, given its far greater genetic divergence in comparison to the other samples, including those obtained from far reaching parts of the globe.
Conclusions
The Gyrocotylidea is a small, but common group of cestodes of holocephalans with a widespread distribution characteristic of a relictual parasite group restricted to a relictual host group. The mostly deep-sea habitat of their hosts represents an unusually stable environment in which this host-parasite system evolved and likely explains their long-term persistence. Other features of the deep-sea, including fjords, are likely to have structured these systems in ways that are not immediately obvious until topography and mechanisms of isolation are considered, and may account for why genetic divergences do not strongly correlate with the degree of geographical separation among samples. Their systematics has been hitherto reliant on morphology and host-associations which in most cases have failed to satisfactorily distinguish species. It is therefore imperative that molecular investigations be employed to guide the circumscription of natural groups. Our results indicate that Gyrocotyle comprises not one cosmopolitan, non-specific species, but a group of distinct, mostly host-specific, species that cannot be distinguished by morphology. Although the recognition of individual lineages and sequence clusters as species is problematical and probably provisional, establishing these conceptions now is justified by the fact that the hosts of the new species are rarely seen; the specimen of the long-nosed chimaera Ha. raleighana (Fig. 7) is the only one RAB has examined in over 30 years of marine trawling, whereas Hy. mirabilis (Fig. 8) is found in numbers, but only at particular depths and localities.
References
Bandoni, S. M., & Brooks, D. R. (1987). Revision and phylogenetic analysis of the Gyrocotylidea Poche, 1926 (Platyhelminthes: Cercomeria: Cercomeromorpha). Canadian Journal of Zoology,65, 2369–2389.
Berland, B., Bristow, G. A., & Grahl Nielsen, O. (1990). Chemotaxonomy of Gyrocotyle (Platyhelminthes: Cercomeria) species, parasites of chimaerid fish (Holocephali), by chemometry of their fatty acids. Marine Biology,105, 185–189.
Bristow, G. A. (1992). On the distribution, ecology and evolution of Gryocotyle [sic] urna, G. confusa and G. nybelini (Cercomeromorpha: Gyrocotylidea) and their host Chimaera monstrosa (Holocephalidea: Chimaeridae) in Norwegian waters, with a review of the species question. Sarsia,77, 119–124.
Bristow, G. A., & Berland, B. (1988). A preliminary electrophoretic investigation of the gyrocotylid parasites of Chimaera monstrosa L. Sarsia,73, 75–77.
Bunkley-Williams, L., & Williams, E. H. (2004). New locality, depth, and size records and species character modifications of some Caribbean deep-reef/shallow slope fishes and a new host and locality record for the chimaera cestodarian. Caribbean Journal of Science,40, 88–119.
Caira, J. N., Jensen, K., & Healy, C. J. (1999). On the phylogenetic relationships among tetraphyllidean, lecanicephalidean and diphyllidean tapeworm genera. Systematic Parasitology,42, 77–151.
Caira, J. N., Jensen, K., Waeschenbach, A., Olson, P. D., & Littlewood, D. T. J. (2014). Orders out of chaos - molecular phylogenetics reveals the complexity of shark and stingray tapeworm relationships. International Journal for Parasitology,44, 55–73.
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution,17, 540–552.
Colin, J. A., Williams, H. H., & Halvorsen, O. (1986). One or three gyrocotylideans (Platyhelminthes) in Chimaera monstrosa (Holocephali)? Journal of Parasitology,72, 10–21.
Gibson, D. I. (1994). Order Gyrocotylidea Poche, 1926. In: Khalil, L. F., Jones, A. & Bray, R. A. (Eds), Keys to the cestode parasites of vertebrates Wallingford: CAB International, pp. 11–13.
Hogans, W. E., & Hurlbut, T. R. (1984). Parasites of the knifenose chimaera, Rhinochimaera atlantica, from the northwest Atlantic Ocean. Canadian Field-Naturalist,98, 365.
ICZN (2012). International Commission on Zoological Nomenclature: Amendment of articles 8, 9, 10, 21 and 78 of the International Code of Zoological Nomenclature to expand and refine methods of publication. Bulletin of Zoological Nomenclature,69, 161–169.
Inoue, J. G., Miya, M., Lam, K., Tay, B.-H., Danks, J. A., Bell, J., et al. (2010). Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): a mitogenomic perspective. Molecular Biology and Evolution,27, 2576–2586.
Karlsbakk, E., Aspholm, P. E., Berg, V., Hareide, N. R., & Berland, B. (2002). Some parasites of the small-eyed rabbitfish, Hydrolagus affinis (Capello, 1867) (Holocephali), caught in deep waters off SW Greenland. Sarsia,87, 179–184.
Katoh, T. (2008). Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics,9, 286–298.
Licht, M., Schmuecker, K., Huelsken, T., Hanel, R., Bartsch, P., & Paeckert, M. (2012). Contribution to the molecular phylogenetic analysis of extant holocephalan fishes (Holocephali, Chimaeriformes). Organisms Diversity & Evolution,12, 421–432.
Littlewood, D. T. J., Curini-Galletti, M., & Herniou, E. A. (2000). The interrelationships of Proseriata (Platyhelminthes: Seriata) tested with molecules and morphology. Molecular Phylogenetics and Evolution,16, 449–466.
Littlewood, D. T. J., & Olson, P. D. (2001). Small subunit rDNA and the Platyhelminthes: Signal, noise, conflict and compromise. In: Littlewood, D. T. J. & Bray, R. A. (Eds), Interrelationships of the Platyhelminthes. London: Taylor & Francis, pp. 262–278.
Maddison, W. P., & Maddison, D. R. (2018). Mesquite: a modular system for evolutionary analysis. Version 3.01. http://www.mesquiteproject.org. Accessed Dec 2019.
Mauchline, J., & Gordon, J. D. M. (1984). Incidence of parasitic worms in stomachs of pelagic and demersal fish of the Rockall Trough, northeastern Atlantic Ocean. Journal of Fish Biology,24, 281–285.
Nylander, J. A. A. (2004). MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. https://www.abc.se/~nylander/. Accessed Dec 2019
Olson, P. D., & Caira, J. N. (1999). Evolution of the major lineages of tapeworms (Platyhelminthes: Cestoidea) inferred from 18S ribosomal DNA and elongation factor-1α. Journal of Parasitology,85, 1134–1159.
Olson, P. D., Littlewood, D. T. J., Bray, R. A., & Mariaux, J. (2001). Interrelationships and evolution of the tapeworms (Platyhelminthes: Cestoda). Molecular Phylogenetics and Evolution,19, 443–467.
Olson, P. D., Poddubnaya, L. G., Littlewood, D. T. J., & Scholz, T. (2008). On the position of Archigetes and its bearing on the early evolution of the tapeworms. Journal of Parasitology,94, 898–904.
Parukhin, A. M. (1966). [On the species composition of the helminth fauna of fishes in the South Atlantic]. Materialy Nauchnoi Konferentsii Vsesoyuznogo Obshchestva Gelʼmintologov, 219–222 (In Russian).
Parukhin, A. M. (1968). Helminthofauna of fishes of South Atlantic. Biologiya Morya, Kiev,14, 96–113. (In Russian).
Ronquist, F., & Huelsenbeck, J. P. (2003). MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics,19, 1572–1574.
Simmons, J. E., Buteau, G. H., Jr., Macinnis, A. J., & Kilejian, A. (1972). Characterization and hybridization of DNAs of gyrocotylidean parasites of chimaeroid fishes. International Journal for Parasitology,2, 273–278.
Talavera, G., & Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology,56, 564–577.
van der Auwera, G., Chapelle, S., & de Wachter, R. (1994). Structure of the large ribosomal subunit RNA of Phytophthora megasperma, and the phylogeny of the oomycetes. FEBS Letters,338, 133–136.
van der Land, J., & Templeman, W. (1968). Two new species of Gyrocotyle (Monogenea) from Hydrolagus affinis (Brito Capello) (Holocephali). Journal of the Fisheries Research Board of Canada,11, 2365–2385.
Waeschenbach, A., Webster, B. L., & Littlewood, D. T. J. (2012). Adding resolution to ordinal level relationships of tapeworms (Platyhelminthes: Cestoda) with large fragments of mtDNA. Molecular Phylogenetics and Evolution,63, 834–847.
Williams, H. H., & Bray, R. A. (1984). Chimaerocestos prudhoei gen. et sp. nov., representing a new family of tetraphyllideans and the first record of strobilate tapeworms from a holocephalan. Parasitology,88, 105–116.
Williams, H. H., Colin, J. A., & Halvorsen, O. (1987). Biology of gyrocotylideans with emphasis on reproduction, population ecology and phylogeny. Parasitology,95, 173–207.
Acknowledgements
RAB is indebted to the crew and officers of the NERC research vessel RRS Discovery, to Professor Imants ‘Monty′ Priede of the Oceanlab Aberdeen, Dr Nigel Merrett and Ms Mary Spencer Jones of the Natural History Museum (NHM), London, and to other colleagues for help with the collection of specimens. We thank Roman Kuchta (Institute of Parasitology, Czech Republic) for bringing accessioning errors in published gyrocotylidean sequences to our attention. We thank the NHM′s Sequencing Unit for assistance in gene sequencing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable institutional, national and international guidelines for the care and use of animals were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article was registered in the Official Register of Zoological Nomenclature (ZooBank) as urn:lsid:zoobank.org:pub:86FBE97B-A44F-48E5-8600-294119A7F304. This article was published as an Online First article on the online publication date shown on this page. The article should be cited by using the doi number. This is the Version of Record.
This article is part of the Topical Collection Cestoda.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bray, R.A., Waeschenbach, A., Littlewood, D.T.J. et al. Molecular circumscription of new species of Gyrocotyle Diesing, 1850 (Cestoda) from deep-sea chimaeriform holocephalans in the North Atlantic. Syst Parasitol 97, 285–296 (2020). https://doi.org/10.1007/s11230-020-09912-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11230-020-09912-w