Abstract
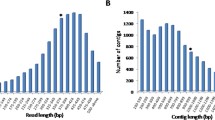
A high-throughput sequencing by 454 pyrosequencing was performed on the transcriptomes of apical meristem tissue of two Jatropha curcas accessions and four jatropha-related species. J. curcas accession (acc.) CN produced 45.61 Mb data from 127,094 reads with 11,579 contigs and 15,645 singletons, while acc. M10 produced 54.52 Mb data from 142,447 reads with 10,964 contigs and 12,069 singletons. J. integerrima acc. KL gave 20.91 Mb from 71,541 reads with 4,551 contigs and 5,144 singletons, while acc. KY gave 48.74 Mb from 149,392 reads with 8,440 contigs and 8,299 singletons. J. multifida (acc. JM) gave 66.72 Mb from 191,654 reads with 17,444 contigs and 13,965 singletons, and J. podagrica (acc. JP) gave 73.28 Mb from 165,228 reads with 14,070 contigs and 16,572 singletons. BLASTX search of all contigs revealed that 1,683 unique proteins were orthologous across all four Jatropha spp. From 432 EST-SSR primer pairs designed and tested on these six accessions, 269 markers showed polymorphism and produced a total of 862 alleles. Based on sequence alignments between J. curcas accessions with low and high phorbol esters (PEs), 20 candidate SNPs were identified in four coding sequences including one gene relating to biosynthesis pathways of PEs. These expressed sequence tag-sample sequence repeat (EST-SSR) markers and candidate single nucleotide polymorphisms (SNPs) will be useful for jatropha breeding in the future.


Similar content being viewed by others
References
Achten WMJ, Verchot L, Franken YJ, Mathijs E, Singh VP, Aerts R, Muys B (2008) Review: Jatropha bio-diesel production and use. Biomass Bioenergy 32:1063–1084
Arreguín JCV, Laclette EI, Moraila BJ, Martínez O, Calzada JPV, Estrella LH, Estrella AH (2009) Deep sampling of the Palomero maize transcriptome by a high throughput strategy of pyrosequencing. BMC Genomics 10:299–308
Benbouza H, Jacquemin JM, Baudoin JP, Mergeai G (2006) Optimization of a reliable, fast, cheap and sensitive silver staining method to detect SSR markers in polyacrylamide gels. Biotechnol Agron Soc Environ 10:77–81
Brooker J (2009) Methods for detoxifying oil seed crops (Patent WO/2010/070264)
Carvalhoa CR, Clarindoa WR, Praca MM, Araújoa FS, Carels N (2008) Genome size, base composition and karyotype of Jatropha curcas L., an important biofuel plant. Plant Sci 174:613–617
Costa GGL, Cardoso KC, Del Bem LEV, Lima AC, Cunha MAS, Camposde LL, Vicentini R, Papes F, Moreira RC, Yunes JA, Campos FAP, Silva MF (2010) Transcriptome analysis of the oil-richseed of the bioenergy crop Jatropha curcas L. BMC Genomics 11:462–470
Deng X, Fang Z, Liu YH (2010) Ultrasonic transesterification of Jatropha curcas L. oil to biodiesel by a two-step process. Energy Convers Manag 51:2802–2807
Deschamps S, Campbell MA (2010) Utilization of next-generation sequencing platforms in plant genomics and genetic variant discovery. Mol Breed 25:553–570
Galun E (2007) Plant patterning: structural and molecular genetic aspects. World Scientific, Singapore
Guo S, Zheng Y, Joung JG, Liu S, Zhang Z, Crasta OR, Sobral BW, Xu Y, Huang S, Fei Z (2010) Transcriptome sequencing and comparative analysis of cucumber flowers with different sex types. BMC Genomics 11:384–396
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Jha TB, Mukherjee P, Datta MM (2007) Somatic embryogenesis in Jatropha curcas Linn., an important biofuel plant. Plant Biotechnol Rep 1:135–140
King AJ, He W, Cuevas JA, Freudenberger M, Ramiaramanana DI, Graham IA (2009) Potential of Jatropha curcas as a source of renewable oil and animal feed. J Exp Bot 60:2897–2905
King AJ, Montes LR, Clarke JG, Affleck J, Li Y, Witsenboer H, van der Vossen E, van der Linde P, Tripathi Y, Tavares E, Shukla P, Rajasekaran T, van Loo EN, Graham IA (2013) Linkage mapping in the oilseed crop Jatropha curcas L. reveals a locus controlling the biosynthesis of phorbol esters which cause seed toxicity. Plant Biotechnol J 11:986–996
King AJ, Li Y, Graham IA (2011) Profiling the developing Jatropha curcas L. seed transcriptome by pyrosequencing. Bioenerg Res 4:211–221
Kumar S, Blaxter ML (2010) Comparing de novo assemblers for 454 transcriptome data. BMC Genomics 11:571–582
Menezes RG, Rao NG, Karanth SS, Kamath A, Manipady S, Pillay VV (2006) Jatropha curcas poisoning. Indian J Pediatr 73:634
Natarajan P, Kanagasabapathy D, Gunadayalan G, Panchalingam J, Shree N, Sugantham PA, Singh KK, Madasamy P (2010) Gene discovery from Jatropha curcas by sequencing of ESTs from normalized and full-length enriched cDNA library from developing seeds. BMC Genomics 11:606–612
Natarajan P, Parani M (2011) De novo assembly and transcriptome analysis of five major tissues of Jatropha curcas L. using GSFLX titanium platform of 454 pyrosequencing. BMC Genomics 12:191–102
Rafalski A (2002) Applications of single nucleotide polymorphisms in crop genetics. Plant Biol 5:94–100
Roth C, Liberles DA (2006) A systematic search for positive selection in higher plants (Embryophytes). BMC Plant Biol 6:12
Saetae D, Kleekayai T, Jayasena V, Suntornsuk W (2011) Functional properties of protein isolate obtained from physic nut (Jatropha curcas L.) seed cake. Food Sci Biotechnol 20:29–37
Sato S, Hirakawa H, Isobe S, Fukai E, Watanabe A, Kato M, Kawashima U, Minami C, Muraki A, Nakazaki N, Takahashi C, Nakayama S, Kishida Y, Kohara M, Yamada M, Tsuruoka H, Sasamoto S, Tabata S, Aizu T, Toyoda A, Shin-I T, Minakuchi Y, Kohara Y, Fujiyama A, Tsuchimoto S, Kajiyama S, Makigano E, Ohmido N, Shibagaki N, Cartagene JA, Wada N, Kohinata T, Atefeh A, Yuasa S, Matsunaga S, Fukui K (2011) Sequence analysis of the genome of an oil bearing tree, Jatropha curcas L. DNA Res 18:65–76
Stukenbrock EH, McDonald BA (2009) Population genetics of fungal and oomycete effectors involved in gene-for-gene interactions. Mol Plant Microbe Interact 22:371–380
Sujatha M (2006) Genetic improvement of Jatropha curcas L.: possibilities and prospects. Indian J Agrofor 8:58–65
Tanya P, Taeprayoon P, Hadkam Y, Srinives P (2011) Genetic diversity among jatropha and jatropha-related species based on ISSR markers. Plant Mol Biol Report 29:252–264
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Wei X, Sujatha M, Aizhong L (2012) Review: genetic diversity in the Jatropha genus and its potential application. CAB Rev 7:1–15
Wicker T, Schlagenhauf E, Graner A, Close TJ, Keller B, Stein N (2006) 454 sequencing put to the test using the complex genome of barley. BMC Genomics 7:275–285
Wen M, Wang H, Xia Z, Zou M, Lu C, Wang W (2010) Development of EST-SSR and genomic-SSR markers to assess genetic diversity in Jatropha curcas L. BMC Res Notes 3:42
Zhang Z, Li J, Zhao XQ, Wang J, Wong KSG, Yu J (2006) KaKs_calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4:259–263
Acknowledgments
This research was supported by a joint Royal Golden Jubilee PhD Scholarship between the Thailand Research Fund and Kasetsart University, the Chair Professor Project of Thailand’s National Science and Technology Development Agency (Grant no. P-11-00599), and the Center of Excellence on Agricultural Biotechnology (AG-BIO/PERDO-CHE), Bangkok, Thailand.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
SNP in scaffold Jcr4S00160 coding for steroid binding proteins. Steroid binding proteins involve with the basic five-carbon unit isopentyldiphosphate (IPP), the initial substrate for PE synthesis (JPEG 533 kb)
Table S1
(DOCX 16 kb)
Table S2
(DOCX 13 kb)
Table S3
(DOCX 13 kb)
Table S4
(DOCX 55 kb)
Table S5
(DOCX 11 kb)
Table S6
(DOCX 14 kb)
Rights and permissions
About this article
Cite this article
Laosatit, K., Tanya, P., Somta, P. et al. De novo Transcriptome Analysis of Apical Meristem of Jatropha spp. Using 454 Pyrosequencing Platform, and Identification of SNP and EST-SSR Markers. Plant Mol Biol Rep 34, 786–793 (2016). https://doi.org/10.1007/s11105-015-0961-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0961-z