Abstract
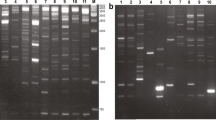
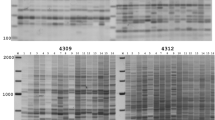
Retrotransposon (RTN)-based markers, such as the inter-retrotransposon amplified polymorphism (IRAP) and the retrotransposon-microsatellite amplified polymorphism (REMAP), are highly informative, multilocus, and reveal insertion polymorphisms among individuals. These markers have been used for evolutionary studies, genetic diversity assessment, DNA fingerprinting, and detection of genetic rearrangements induced by allopolyploidization. The hexaploid tritordeum (HchHchAABB; 2n = 6x = 42) is an allopolyploid produced from crosses between wild barley (Hordeum chilense Roem. et Schultz.) (HchHch; 2n = 2x = 14) and durum wheat (Triticum turgidum L. conv. durum) (AABB; 2n = 4x = 28). With this study, we carried out the DNA fingerprinting of two newly formed hexaploid tritordeum lines (HT22 and HT27) and their respective parents, line H1 of H. chilense and line T81 of durum wheat, based on IRAPs, REMAPs and inter-simple sequence repeats (ISSRs), in order to detect potential rearrangements in tritordeum derived from polyploidization. The amphiploid nature of the HT22 and HT27 individuals was successfully confirmed after fluorescence in situ hybridization (FISH), which was performed on their mitotic chromosome spreads with genomic DNA from H. chilense and 45S ribosomal DNA (rDNA), simultaneously, as probes. Six combinations of LTR (long terminal repeat) primers and seven combinations of one LTR and one SSR (simple sequence repeat) primers successfully produced IRAPs and REMAPs, respectively, in both tritordeum lines, and their respective parents. ISSRs were produced with three SSR primers (8081, 8082, and 8564). The analysis of the presence/absence of bands among the tritordeum lines and the respective parents allowed the detection of polymorphic bands: (1) shared by tritordeum and one of the parents; (2) exclusively amplified in tritordeum; and (3) exclusively present in one of the parents. Once no polymorphism was detected among the individuals of each parental species, the polymorphic bands that fit into the second and third cases probably constituted rearrangements in the newly formed tritordeums that arose in response to allopolyploidization, which resulted from the loss of parental bands or, conversely, from the appearance of novel bands not seen in the parental species. Most of the polymorphic IRAPs in tritordeum were shared with the female parent (H. chilense), while most of the polymorphic REMAPs and ISSRs were common to the male parent (durum wheat), but globally, most of the bands inherited by tritordeum had a wheat origin. In conclusion, these dominant markers were successful for DNA fingerprinting and detection of rearrangements in newly formed tritordeum derived from responses to allopolyploidization.



Similar content being viewed by others
References
Alavi-Kia SS, Mohammadi SA, Aharizad S, Moghaddam M (2008) Analysis of genetic diversity and phylogenetic relationships in Crocus genus of Iran using inter-retrotransposon amplified polymorphism. Biotechnol Biotechnol EQ 22(2008/3):795–800
Baumel A, Ainouche M, Kalendar R, Schulman AH (2002) Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C.E. Hubbard (Poaceae). Mol Biol Evol 19(8):1218–1227
Bento M, Pereira HS, Rocheta M, Gustafson P, Viegas W, Silva M (2008) Polyploidization as a retraction force in plant genome evolution: sequence rearrangements in Triticale. PLoS ONE 3:e1402. doi:10.1371/journal.pone.0001402
Bento M, Gustafson P, Viegas W, Silva M (2010) Genome merger: from sequence rearrangements in triticale to their elimination in wheat–rye addition lines. Theor Appl Genet 121:489–497
Branco CJS, Vieira EA, Malone G, Kopp MM, Malone E, Bernardes A, Mistura CC, Carvalho FIF, Oliveira CA (2007) IRAP and REMAP assessments of genetic similarity in rice. J Appl Genet 48(2):107–113
Cabo S, Carvalho A, Martín A, Lima-Brito J (2013) Structural rearrangements detected in newly-formed hexaploid tritordeum after three sequential FISH experiments with repetitive DNA sequences. J Genet (in press)
Calonje M, Martín-Bravo S, Dobes C, Gong W, Jordon-Thaden I, Kiefer C, Kiefer M, Paule J, Schmickl R, Koch MA (2009) Non-coding nuclear DNA markers in phylogenetic reconstruction. Plant Syst Evol 282:257–280
Carvalho AIF (2004) Emparelhamento meiótico e DNA fingerprint em anfiplóides e híbridos interespecíficos da Tribo Triticeae. Master Thesis in Genetic Resources and Breeding of Forestry and Crop species, University of Tras-os-Montes and Alto Douro, Vila Real, Portugal
Carvalho A, Matos M, Lima-Brito J, Guedes-Pinto H, Benito C (2005) DNA fingerprint of F1 interspecific hybrids from the Triticeae tribe using ISSRs. Euphytica 143:93–99
Carvalho A, Guedes-Pinto H, Heslop-Harrison JS, Lima-Brito J (2008) Wheat neocentromeres found in F1 triticale × tritordeum hybrids (AABBRHch) after 5-azacytidine treatment. Plant Mol Biol Report 26:46–52
Carvalho A, Martín A, Heslop-Harrison JS, Guedes-Pinto H, Lima-Brito J (2009) Identification of the spontaneous 7BS/7RL intergenomic translocation in one F1 multigeneric hybrid from the Triticeae tribe. Plant Breed 128(1):105–108
Carvalho A, Guedes-Pinto H, Martins-Lopes P, Lima-Brito J (2010) Genetic variability analysis in Old Portuguese bread wheat cultivars assayed by IRAP and REMAP markers. Ann Appl Biol 156(3):337–345
Carvalho A, Guedes-Pinto H, Lima-Brito J (2012) Genetic diversity in Old Portuguese durum wheat cultivars assessed by retrotransposon-based markers. Plant Mol Biol Report 30:578–589
Castillo A, Ramírez MC, Martín AC, Kilian A, Martín A, Atienza SG (2013) High-throughput genotyping of wheat-barley amphiploids utilising diversity array technology (DArT). BMC Plant Biol 13:87, http://www.biomedcentral.com/1471-2229/13/87
Chadha S, Gopalakrishna T (2005) Retrotransposon-microsatellite amplified polymorphism (REMAP) markers for genetic diversity assessment of the rice blast pathogen (Magnaporthe grisea). Genome 48:943–945
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885
Jones RN, Hegarty M (2009) Order out of chaos in the hybrid plant nucleus. Cytogenet Genome Res 126(4):376–389
Kalendar R, Schulman AH (2006) IRAP and REMAP for retrotransposon-based genotyping and fingerprinting. Nat Protoc 1(5):2478–2484
Kalendar R, Grob T, Regina M, Suoniemi A, Schulman A (1999) IRAP and REMAP: two retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet 98:704–711
Kashkush K, Feldman M, Levy AA (2002) Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160:1651–1659
Kraitshtein Z, Yaakov B, Khasdan V, Kashkush K (2010) Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics 186:801–812
Leigh F, Kalendar R, Lea V, Lee D, Donini P, Schulman AH (2003) Comparison of the utility of barley retrotransposon families for genetic analysis by molecular marker techniques. Mol Gen Genomics 269:464–474
Levy AA, Feldman M (2002) The impact of polyploidy on grass genome evolution. Plant Physiol 130:1587–1593
Lima-Brito J, Guedes-Pinto H, Heslop-Harrison JS (1998) The activity of nucleolar organizing chromosomes in multigeneric F1 hybrids involving wheat, triticale, and tritordeum. Genome 41:763–768
Lima-Brito J, Carvalho A, Martín A, Heslop-Harrison JS, Guedes-Pinto H (2006) Morphological, yield, cytological and molecular characterisation of a bread wheat x tritordeum F1 hybrid. J Genet 85:123–131
Ma XF, Gustafson JP (2005) Genome evolution of allopolyploids: a process of cytological and genetic diploidization. Cytogenet Genome Res 109(1–3):236–249
Manninen O, Kalendar R, Robinson J, Schulman AH (2000) Application of BARE-1 retrotransposon markers to the mapping of a major resistance gene for net blotch in barley. Mol Gen Genet 264:325–334
Martín A, Sánchez-Monge Laguna E (1982) Cytology and morphology of the amphiploid Hordeum chilense x Triticum turgidum conv. durum. Euphytica 31:262–267
Ozkan H, Levy AA, Feldman M (2001) Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13:1735–1747
Queen RA, Gribbon M, James C, Jack P, Flavell AJ (2004) Retrotransposon-based molecular markers for linkage and genetic diversity analysis in wheat. Mol Gen Genomics 271:91–97
Sabot F, Schulman AH (2007) Template switching can create complex LTR retrotransposon insertions in Triticeae genomes. BMC Genomics 8:247. doi:10.1186/1471-2164-8-247
Sabot F, Guyot R, Wicker T, Chantret N, Laubin B, Chalhoub B, Leroy P, Sourdille P, Bernard M (2005) Updating of transposable element annotations from large wheat genomic sequences reveals diverse activities and gene associations. Mol Gen Genomics 274:119–130
Saeidi H, Rahiminejad MR, Heslop-Harrison JS (2008) Retroelement insertional polymorphisms, diversity and phylogeography within diploid, D-genome Aegilops tauschii (Triticeae, Poaceae) sub-taxa in Iran. Ann Bot 101:855–861
Wegscheider E, Benjak A, Forneck A (2009) Clonal variation in Pinot noir revealed by S-SAP involving universal retrotransposon-based sequences. Am J Enol Vitic 60(1):104–109
Acknowledgments
This work was partially supported by the HY-WHEAT project - International Consortium (P-KBBE/AGR-GPL/0002/2010), funded by the COMPETE program, QREN, EU, and the Portuguese Foundation for Science and the Technology (FCT). 
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cabo, S., Carvalho, A., Rocha, L. et al. IRAP, REMAP and ISSR Fingerprinting in Newly Formed Hexaploid Tritordeum (X Tritordeum Ascherson et Graebner) and Respective Parental Species. Plant Mol Biol Rep 32, 761–770 (2014). https://doi.org/10.1007/s11105-013-0684-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-013-0684-y