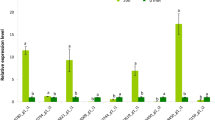
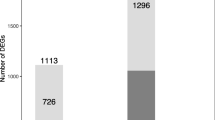
Abstract
Hibiscus tiliaceus, a mangrove associate, is an ideal plant for studying salt tolerance and adaptation since it can inhabit both inland and littoral habitats. In this study, we explored the expression profiles of H. tiliaceus under salt stress using a full-length cDNA microarray. Four hundred eighty-six salt-responsive unigenes were identified in H. tiliaceus; 224 of which had high sequence similarity to Arabidopsis. Many genes identified are known to be salt-stress responsive. Furthermore we examined the physiological performance of H. tiliaceus under salt stress. Physiological analysis displayed decrease in ratio of K+/Na+ and negative influence on photosynthesis of H. tiliaceus. Our study indicated that to survive under high salinine intertidal environments, H. tiliaceus evolved its own mechanisms to re-gain both ionic and osmotic homeostasis through coordinated engagement of genes associated with gene transcription, signaling, and down-stream cell transport and detoxification pathways.




Similar content being viewed by others
References
Ambard-Bretteville F, Sorin C, Rebeille F et al (2003) Repression of formate dehydrogenase in Solanum tuberosum increases steady-state levels of formate and accelerates the accumulation of proline in response to osmotic stress. Plant Mol Bio 52:1153–1168
Apse MP, Blumwald E (2002) Engineering salt tolerance in plants. Curr Opin Biotechnol 13:146–150
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by Overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285:1256–1258
Badejo AA, Fujikawa Y, Esaka M (2009) Gene expression of ascorbic acid biosynthesis related enzymes of the Smirnoff-Wheeler pathway in acerola (Malpighia glabra). J Plant Physiol 166:652–660
Blumwald E (2000) Sodium transport and salt tolerance in plants. Curr Opin Cell Bio 12:431–434
Chao DY, Luo YH, Shi M et al (2005) Salt-responsive genes in rice revealed by cDNA microarray analysis. Cell Res 15:796–810
Cramer GR, AE JG et al (2007) Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct Integr Genomics 7:111–134
Dat J, Vandenabeele S, Vranová E et al (2000) Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci 57:779–795
Deveaux Y, Toffano-Nioche C, Claisse G et al (2008) Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol Bio 8:291
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95:14863–14868
FAO (2008) FAO land and plant nutrition management service. Available at: http://www.fao.org/ag/agl/agll/spush. Accessed 25 April
Fu X, Huang Y, Deng S et al (2005) Construction of a SSH library of Aegiceras corniculatum under salt stress and expression analysis of four transcripts. Plant Sci 169:147–154
Gao C, Wang Y, Liu G et al (2010) Cloning of ten Peroxidase (POD) genes from Tamarix Hispida and characterization of their responses to Abiotic stress. Plant Mol Biol Rep 28:77–89
Hans-Hubert K, Dorothea B, Wei Y et al (2004) The ALDH gene superfamily of Arabidopsis. Trends Plant Sci 9(8):371–377
Harmer SL, Hogenesch JB, Straume M et al (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290:2110–2113
Hasegawa PM, Bressan RA, Zhu JK et al (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Bio 51:463–499
Ishitani M, Majumder AL, Bornhouser A et al (1996) Coordinate transcriptional induction of myo-insitol metabolism during environmental stress. Plant J 9:537–548
Jiang Y, Deyholos MK (2006) Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol 6:25
Jithesh MN, Prashanth SR, Sivaprakash KR et al (2006) Antioxidative response mechanisms in halophytes: their role in stress defence. J Genet 85(3):237–254
Kawasaki S, Borchert C, Deyholos M et al (2001) Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13:889–906
Kim MJ, Lim GH, Kim ES et al (2007) Abiotic and biotic stress tolerance in Arabidopsis overexpressing the Multiprotein bridging factor 1a (MBF1a) transcriptional coactivator gene. Biochem Biophys Res Commun 354(2):440–446
Kreps JA, Wu Y, Chang HS et al (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130:2129–2141
Lee CE, Mitchell-Olds T (2006) Preface to the special issue: ecological and evolutionary genomics of populations in nature. Mol Ecol 15:1193–1196
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−⊿⊿Ct method. Methods 25:402–408
Ma S, Gong Q, Bohnert HJ (2006) Dissecting salt stress pathways. J Exp Bot 57:1097–1107
Mittler R, Vanderauwera S, Gollery M et al (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9(10):490–498
Miyama M, Hanagata N (2007) Microarray analysis of 7029 gene expression patterns in burma mangrove under high-salinity stress. Plant Sci 172(5):948–957
Parida AK, Das AB, Mohanty P (2004) Investigations on the anti-oxidative defence responses to NaCl stress in a mangrove, Bruguiera parviflora: differential regulations of isoforms of some antioxidative enzymes. Plant Growth Regul 42:213–226
Popp M, Larher F, Weigel P (1985) Osmotic adaptation in Australia mangroves. Plant Ecol 61:247–254
Ren ZH, Gao JP, Li LG et al (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Gent 37(10):1141–1146
Rensink WA, Iobst S, Hart A et al (2005) Gene expression profiling of potato responses to cold, heat, and salt stress. Funct Integr Genomics 5:201–207
Sahi C, Singh A, Blumwald E et al (2006) Beyond osmolytes and transporters: novel plant salt-stress tolerance-related genes from transcriptional profiling data. Physiologia Plantarum 127:1–9
Schaffer R, Landgraf J, Accerbi M et al (2001) Microarray analysis of diurnal and circadianregulated genes in Arabidopsis. Plant Cell 13:113–123
Shigeoka S, Ishikawa T, Tamoi M et al (2002) Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot 53(372):1305–1319
Taji T, Seki M, Satou M et al (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol 135:1697–1709
Tang R, Li C, Xu K et al (2010) Isolation, functional characterization, and expression pattern of a vacuolar Na+/H+ Antiporter gene TrNHX1 from Trifolium repens L. Plant Mol Biol Rep 28:102–111
Tomlinson PB (1986) The botany of mangroves. Cambridge University, Cambridge
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci 98(9):5116–5121
Wang H, Miyazaki S, Kawai K et al (2003a) Temporal progression of gene expression responses to salt shock in maize roots. Plant Mol Bio 52:873–891
Wang WX, Vinocur B, Altman A (2003b) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14
Wang B, Davenport RJ, Volkov V et al (2006) Low unidirectional sodium influx into root cells restricts net sodium accumulation in Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana. J Exp Bot 57:1161–1170
Wang X, Dong J, Liu Y et al (2010) A novel dehydration-responsive element-binding protein from Caragana korshinskii is involved in the response to multiple abiotic stresses and enhances stress tolerance in transgenic tobacco. Plant Mol Biol Rep 28:664–675
Wu C, Gao X, Kong X et al (2009) Molecular cloning and functional analysis of a Na+/H+ Antiporter gene ThNHX1 from a halophytic plant Thellungiella halophila. Plant Mol Biol Rep 27:1–12
Yan S, Tang Z, Su W et al (2005) Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics 5:235–244
Yang G, Zhou R, Tang T et al (2008) Simple and efficient isolation of high-quality total RNA from Hibiscus tiliaceus, a mangrove associate and its relatives. Prep Biochem Biotechnol 38:257–264
Yeo AR, Lee K-S, Izard P et al (1991) Short- and long-term effects of salinity on leaf growth in rice (Oryza sativa L.). J Exp Bot 42:881–889
Yokoi S, Quintero FJ, Cubero B et al (2002) Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J 30:529–539
Zhang J, Deng Z, Cao S et al (2008) Isolation of six novel Aquaporin genes from Triticum aestivum L. and functional analysis of TaAQP6 in water redistribution. Plant Mol Biol Rep 26:32–45
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 16:66–67
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Acknowledgments
We thank Jiajun Zhang, Suisui Dong, Yan Li, Guorong Wen, Haijun Wen, Jin Liu, Wei Wu, Linghui Wu, Shulin Deng, and San Liang for their help in the experiment. Special appreciation should be given to Qiang Fan for his helping in material collection. This study is supported by grants from the National Natural Science Foundation of China (30730008, 40976081, 30800060, 30970208, and 40876075), the National Basic Research Program of China (2007CB815701), National S&T Major Project of China (2009ZX08010-017B), the Natural Science Foundation of Guangdong Province (8451027501001492, 8151027501000089), Ministry of Education Foundation of China (20070558030), the Fundamental Research Funds for the Central Universities (09lgpy35, 10lgpy20) and the Visiting Scholar Foundation of State Key Lab of Biocontrol (SKLBC09F06).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, G., Zhou, R., Tang, T. et al. Gene Expression Profiles in Response to Salt Stress in Hibiscus Tiliaceus . Plant Mol Biol Rep 29, 609–617 (2011). https://doi.org/10.1007/s11105-010-0267-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-010-0267-0