Abstract
Key message
We provided a chromosome-length assembly of B. nigra and show the comprehensive chromosome-scale variations among Brassica genomes.
Abstract
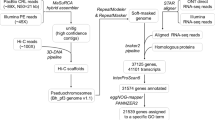
Chromosome-level assembly of the Brassica species, which include many important crops, is essential for the agricultural and evolutionary studies. While the present B. nigra chromosomes was connected with genetic map of B. juncea, hindering the comparative analysis of the B chromosomes. Here we present a chromosome-length B. nigra assembly constructed with Hi-C connections and its variations on chromosome level compared with other Brassica species. We produced an assembly of 484 Mb annotated with 51,829 genes, of which 393 Mb were anchored onto 8 chromosomes, taking 81.26% of the assembly. Comparison of the B chromosomes shows high concordance of the two B. nigra assemblies and reveals comprehensive variations of the B chromosomes after polyploidization and gene loss in syntenic regions. Chromosome blocks with variations have lower gene density and higher TE content. Furthermore, we compared the chromosomes of the three major Brassica diploids, which showed that most of the variations between B and A/C had completed before A/C divergence and there are more variations on C chromosomes after their divergence. In summary, our work presents a chromosome-length assembly of B. nigra and comprehensive comparative analysis of the Brassica chromosomes, which provides a useful reference for other studies and comprehensive information of Brassica chromosome evolution.




Similar content being viewed by others
References
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815. https://doi.org/10.1038/35048692
Bao W, Kojima KK, Kohany O (2015) Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6:11. https://doi.org/10.1186/s13100-015-0041-9
Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94. https://doi.org/10.1006/jmbi.1997.0951
Burton JN, Adey A, Patwardhan RP, Qiu R, Kitzman JO, Shendure J (2013) Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. https://doi.org/10.1038/nbt.2727
Chalhoub B, Denoeud F, Liu S, Parkin I, a. P, Tang H, Wang X et al (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345(6199):950–953. https://doi.org/10.1126/science.1253435
Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC et al (2017) De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356(6333):92–95. https://doi.org/10.1126/science.aal3327
Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES et al (2016a) Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst 3:99–101. https://doi.org/10.1016/j.cels.2015.07.012
Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES et al (2016b) Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3:95–98. https://doi.org/10.1016/j.cels.2016.07.002
Ellinghaus D, Kurtz S, Willhoeft U (2008) LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform 9(1):18. https://doi.org/10.1186/1471-2105-9-18
Elsik CG, Mackey AJ, Reese JT, Milshina NV, Roos DS, Weinstock GM (2007) Creating a honey bee consensus gene set. Genome Biol 8:R13. https://doi.org/10.1186/gb-2007-8-1-r13
Ermolaeva MD, Wu M, Eisen JA, Salzberg SL (2003) The age of the Arabidopsis thaliana genome duplication. Plant Mol Biol 51:859–866. https://doi.org/10.1023/A:1023001130337
Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ et al (2017) InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res 45(D1):D190–D199. https://doi.org/10.1093/nar/gkw1107
Gaeta RT, Chris Pires J (2010) Homoeologous recombination in allopolyploids: The polyploid ratchet. New Phytol 186:18–28. https://doi.org/10.1111/j.1469-8137.2009.03089.x
Han Y, Wessler SR (2010) MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res 38(22):e199–e199. https://doi.org/10.1093/nar/gkq862
Hu TT, Pattyn P, Bakker EG, Cao J, Cheng J, Richard M et al (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43:476–481. https://doi.org/10.1038/ng.807
Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A et al (2009) High-throughput genotyping by whole-genome resequencing. Genome Res 19(6):1068–1076. https://doi.org/10.1101/gr.089516.108
Hunt M, Kikuchi T, Sanders M et al (2013) REAPR: a universal tool for genome assembly evaluation. Genome Biol 14(5):R47. https://doi.org/10.1186/gb-2013-14-5-r47
Johnston JS, Pepper AE, Hall AE, Chen ZJ, Hodnett G, Drabek J, Lopez R, Price HJ (2005) Evolution of genome size in Brassicaceae. Ann Bot 95(1):229–235. https://doi.org/10.1093/aob/mci016
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C et al (2014) InterProScan 5: Genome-scale protein function classification. Bioinformatics. https://doi.org/10.1093/bioinformatics/btu031
Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M et al (2014) Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res 24:1384–1395. https://doi.org/10.1101/gr.170720.113
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44(D1):D457–D462. https://doi.org/10.1093/nar/gkv1070
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664. https://doi.org/10.1101/gr.229202
Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC (2011) Adaptive seeds tame genomic sequence comparison. Genome Res 21(3):487–493. https://doi.org/10.1101/gr.113985.110
Korf I (2004) Gene finding in novel genomes. BMC Bioinform 5(1):59. https://doi.org/10.1186/1471-2105-5-59
Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, Austin MD et al (2012) Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol 30(8):771. https://doi.org/10.1038/nbt.2303
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Lieberman-aiden E, Berkum NL, Van Williams L, Imakaev M, Ragoczy T, Telling A et al (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326(5950):289–294. https://doi.org/10.1126/science.1178746
Liu S, Yeh CT, Ji T, Ying K, Wu H, Tang HM et al (2009) Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet 5(11):e1000733. https://doi.org/10.1371/journal.pgen.1000733
Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IAP et al (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5:3930. https://doi.org/10.1038/ncomms4930
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J et al (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. https://doi.org/10.1186/2047-217X-1-18
Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassiceae. Genome Res 15:516–525. https://doi.org/10.1101/gr.3531105
Majoros WH, Pertea M, Salzberg SL (2004) TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20(16):2878–2879. https://doi.org/10.1093/bioinformatics/bth315
Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A (2018) MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol 14(1):e1005944. https://doi.org/10.1371/journal.pcbi.1005944
Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T et al (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544(7651):427. https://doi.org/10.1038/nature22043
Navabi ZK, Parkin IAP, Pires JC, Xiong Z, Thiagarajah MR, Gooda G et al (2010) Introgression of B-genome chromosomes in a doubled haploid population of Brassica napus × B. carinata. Genome 53(8):619–629. https://doi.org/10.1139/g10-039.Introgression
Navabi ZK, Stead KE, Pires JC, Xiong Z, Sharpe AG, Parkin IAP et al (2011) Analysis of B-genome chromosome introgression in interspecific hybrids of Brassica napus × B. carinata. Genetics 187(3):659–673. https://doi.org/10.1534/genetics.110.124925
Navabi ZK, Huebert T, Sharpe AG, O’Neill CM, Bancroft I, Parkin IAP (2013) Conserved microstructure of the Brassica B genome of Brassica nigra in relation to homologous regions of Arabidopsis thaliana, B. rapa and B. oleracea. BMC Genom 14(1):250. https://doi.org/10.1186/1471-2164-14-250
Paape T, Zhou P, Branca A, Briskine R, Young N, Tiffin P (2012) Fine-scale population recombination rates, hotspots, and correlates of recombination in the Medicago truncatula genome. Genome Biol Evol 4(5):726–737. https://doi.org/10.1093/gbe/evs046
Parkin IAP, Koh C, Tang H et al (2014) Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol 15(6):R77. https://doi.org/10.1186/gb-2014-15-6-r77
Salamov AA, Solovyev VV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10(4):516–522. https://doi.org/10.1101/gr.10.4.516
Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP et al (2015) HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol 16(1):259. https://doi.org/10.1186/s13059-015-0831-x
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. https://doi.org/10.1093/bioinformatics/btv351
Sirén J, Välimäki N, Mäkinen V (2014) HISAT2—fast and sensitive alignment against general human population. IEEE/ACM Trans Comput Biol Bioinform 11:375–388. https://doi.org/10.1109/TCBB.2013.2297101
Soltis DE, Visger CJ, Blaine Marchant D, Soltis PS (2016) Polyploidy: Pitfalls and paths to a paradigm. Am J Bot 103:1146–1166. https://doi.org/10.3732/ajb.1500501
Stanke M, Steinkamp R, Waack S, Morgenstern B (2004) AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res 32:W309–W312. https://doi.org/10.1093/nar/gkh379
Tang H, Lyons E, Pedersen B, Schnable JC, Paterson AH, Freeling M (2011) Screening synteny blocks in pairwise genome comparisons through integer programming. BMC Bioinform 12:102. https://doi.org/10.1186/1471-2105-12-102
Tarailo-Graovac M, Chen N (2009) Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinform. https://doi.org/10.1002/0471250953.bi0410s25
Tian Z, Rizzon C, Du J, Zhu L, Bennetzen JL, Jackson SA et al (2009) Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons? Genome Res 19(12):2221–2230. https://doi.org/10.1101/gr.083899.108
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR et al (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. https://doi.org/10.1038/nprot.2012.016
UniProt Consortium T (2018) UniProt: the universal protein knowledgebase. Nucleic Acids Res 46(5):2699–2699. https://doi.org/10.1093/nar/gky092
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S et al (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039. https://doi.org/10.1038/ng.919
Wen X, Zhong S (2018) 3D genome. University of California. ISBN:987-1-17325643-0-5
Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249. https://doi.org/10.1023/A:1006392424384
Yang J, Liu D, Wang X, Ji C, Cheng F, Liu B et al (2016) The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat Genet 48:1225–1232. https://doi.org/10.1038/ng.3657
Zhang L, Cai X, Wu J, Liu M, Grob S, Cheng F et al (2018) Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic Res 5(1):50. doi.https://doi.org/10.1038/s41438-018-0071-9
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31570539, 31370258, 31601042) and Basic Research Program Support by Shenzhen Municipal Government (JCYJ20151015162041454 to W. C. and JCYJ20150529150505656 to X. Liu).
Author information
Authors and Affiliations
Contributions
Jianbo Wang and Xin Liu are the principle investigators and designed the project. Xiaoming Wu provided the samples sequenced in this research. Wenbin Chen, Qiwu Xu, Guangyi Fan extracted the DNA and RNA. Qiwu Xu and Guangyi Fan did the Hi-C experiment. Wenliang Wang did the genomic assembly, Hi-C data analysis, genome annotation and chromosome comparative analysis. Xing Liu and Haorui Zhang performed quality control analysis of the assembly. Rui Guan performed transcriptome analysis. Wenliang Wang wrote this manuscript. Bo Song, Jianbo Wang and Wenbin Chen edited the manuscript. All authors read and commented on the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Accession number
The genome and transcriptome data from this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/Traces/sra/) and added to the study number SRP074393.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, W., Guan, R., Liu, X. et al. Chromosome level comparative analysis of Brassica genomes. Plant Mol Biol 99, 237–249 (2019). https://doi.org/10.1007/s11103-018-0814-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-018-0814-x