Abstract
Purpose
Previously, we have shown that complexes of plasmid DNA with the biodegradable polymer poly(2-dimethylamino ethylamino)phosphazene (p(DMAEA)-ppz) mediated tumor selective gene expression after intravenous administration in mice. In this study, we investigated the effect of p(DMAEA)-ppz molecular weight on both in vitro and in vivo tumor transfection, as well as on complex induced toxicity.
Materials and Methods
p(DMAEA)-ppz with a broad molar mass distribution was fractionated by preparative size exclusion chromatography. Polyplexes consisting of plasmid DNA and the collected polymer fractions were tested for biophysical properties, (cyto)toxicity and transfection activity.
Results
Four p(DMAEA)-ppz fractions were collected with weight average molecular weights ranging from 130 to 950 kDa, and with narrow molecular mass distributions (Mw/Mn from 1.1 to 1.3). At polymer-to-DNA (N/P) ratios above 6, polyplexes based on these polymers were all positively charged (zeta potential 25–29 mV), and had a size of 80–90 nm. The in vitro cytotoxicity of the polyplexes positively correlated with polymer molecular weight. The in vitro transfection activity of the different polyplexes depended on their N/P ratio, and was affected by the degree of cytotoxicity, as well as the colloidal stability of the different polyplexes. Intravenous administration of polyplexes based on the high molecular weight polymers led to apparent toxicity, as a result of polyplex-induced erythrocyte aggregation. On the other hand, administration of polyplexes based on low molecular weight p(DMAEA)-ppz’s (Mw 130 kDa) did not show signs of toxicity and resulted in tumor selective gene expression.
Conclusion
Polymer molecular weight fractionation enabled us to optimize the transfection efficiency/toxicity ratio of p(DMAEA)-ppz polyplexes for in vitro and in vivo tumor transfection.
Similar content being viewed by others
INTRODUCTION
Non-viral gene delivery holds promise as a therapy against cancer. The genes used for application in anticancer gene medicine encode for antigens, cytokines and suicide proteins (1). In order to efficiently reach metastatic tumors, intravenous administration is the preferred route of administration. In this light, cationic polymers can be useful. The condensation of plasmid DNA using cationic polymers results in positively charged, nanosized polyplexes with improved resistance against nucleases present in blood and other extracellular fluids. In addition, positively charged polyplexes are readily taken up by the target cells. Moreover, surface modification of polyplexes enables improved circulation kinetics and tumor targeting. Indeed, the application of polymers for cancer gene therapy so far has resulted in tumor regression in several animal models (1,2).
Cationic polymers currently used for gene delivery are mostly non-degradable and cytotoxic, limiting their widespread use. The application of biodegradable polymers in polyplexes is a means to prevent long-term polymer tissue accumulation and therefore limit toxicity (3). Ideally, the degradation of the polymer will result in low molecular weight fragments which will be subjected to glomerular kidney filtration or will be metabolized (4). Recently, we reported on the application of the biodegradable poly(2-dimethylamino ethylamino)phosphazene (p(DMAEA)-ppz) for tumor gene delivery (Fig. 1). It was shown that polyplexes based on p(DMAEA)-ppz mediated gene expression to a similar extent as linear polyethylenimine (PEI). Gene expression was confined to the tumor tissue, whereas the use of PEI polyplexes also resulted in substantial expression in non-target organs, particularly in lungs, spleen and liver (5).
Biodegradable as well as non-biodegradable gene delivery polymers generally have a broad molecular weight distribution. Several studies have reported on the importance of polymer molecular weight in the outcome of transfection (6–17). From these studies, it appears that polymer molecular weight not only affects the transfection efficiency of the polyplexes, but also polyplex formation and cytotoxicity. Moreover, polymer molecular weight was found to affect the pharmacokinetics of the polyplexes (9,13,14). For a better understanding of how polymer molecular weight affects transfection activity, these studies are of limited use as the polymers were poorly characterized, had a broad molecular weight distribution, or varied in additional characteristics like branching and conjugation with shielding or targeting ligands. Moreover, several studies included only two different molecular weights, or focused on parameters other than transfection activity. To our knowledge, only few studies have related polymer molecular weight and in vitro transfection activity, using a series of polymers with narrow molecular weight distributions (18–21). None of these studies addressed the additional issue of intravenous administration and/or in vivo tumor transfection.
In this study, we used size exclusion chromatography to fractionate a batch of p(DMAEA)-ppz with a broad molecular weight distribution. We obtained four fractions with different molecular weights and relatively narrow molecular weight distributions. Using these fractions, we were able to assess the effect of polymer molecular weight on the transfection efficiency/toxicity ratio of p(DMAEA)-ppz polyplexes for in vitro and in vivo tumor transfection.
MATERIALS AND METHODS
Materials
The plasmid used in this study, pcDNA3Luc, consists of the coding region for firefly luciferase inserted in the plasmid pcDNA3 (Invitrogen, UK) and is under the transcriptional control of the cytomegalovirus immediate promoter. Endotoxin-free pcDNA3Luc was produced by Plasmid Factory (Bielefeld, Germany). An ethidium bromide (EtBr) stock solution was obtained from Sigma (Zwijndrecht, The Netherlands). Luciferase assay reagent, reporter gene lysis buffer, and Quantilum recombinant luciferase were obtained from Promega (Leiden, The Netherlands).
Polymer Synthesis
Poly(2-dimethylamino ethylamino)phosphazene (p(DMAEA)-ppz) was synthesized and purified as previously described (22,23). In brief, poly(dichloro)phosphazene was synthesized from hexachlorocyclotriphosphazene at 250°C in 1,2,4, trichlorobenzene using sulfamic acid and CaSO42H2O as an initiating system (24). Next, the formed poly(dichlorophosphazene) was substituted with 2-dimethylamino ethylamine. The resulting polymer was purified by extensive dialysis against water and THF, and the solvent was removed under reduced pressure. Finally, the polymer was dissolved in water and collected after freeze-drying.
Preparative SEC Fractionation
A batch of the starting polymer p(DMAEA)-ppz with a broad molar mass distribution was fractionated using a size exclusion chromatography (SEC) system. The SEC system consisted of a Waters 600 EF pump, connected to a Waters 2700 sample manager. A semi-preparative column set (Shodex OHpak SB-LG, OHpak SB-2006 M) was used. The eluent was 0.3 M NaAc (pH 5.0) with a flow rate of 1.5 ml/min. This eluent suppresses ionic interactions between the polymer and the stationary phase (25).
For each fractionation run, 500 μl of solution of p(DMAEA)-ppz (10 mg/ml in eluent) was injected onto the column. Ten fractions were collected at 2 min intervals. The corresponding fractions from 400 repeated runs were pooled, extensively dialyzed against water and collected after freeze-drying. From the resulting ten fractions, two high molecular weight fractions were selected (ppz950 and ppz570) and used for experimental evaluations. Two fractions with intermediate molecular weight were dissolved, pooled and subjected to a second fractionation, using the same SEC settings as described above. To this end, five fractions were collected at 3 min intervals, dialyzed and freeze-dried. From the resulting fractions, two low molecular weight fractions (ppz290 and ppz130) were selected for further evaluations.
Characterization of the Polymer Fractions
The average molecular weights and the molecular weight distribution of the fractionated polymers were determined by SEC, using a Viscotek VE 2001 system (25). Two Shodex SB-804 M columns with a pre-column (Shodex OH-pak SB-G) were connected to a Triple Detector Array 302, equipped with a low and a right angle light scattering detector, a viscometer detector and a refractive index detector. The eluent was 0.3 M NaAc (pH 4.4) with a flow-rate of 1.0 ml/min. The refractive index increment (dn/dc) of p(DMAEA)-ppz was determined by injecting p(DMAEA)-ppz solutions with different concentrations (1–10 mg/ml) directly into the refractive index detector. Molecular weight data analysis was performed by OmniSEC 4.1 software.
An 31P-Nuclear magnetic resonance (NMR) spectrum of the fractionated polymers was recorded on a Varian Inova 500 MHz spectrometer (Varian, USA) (22).
Preparation of the Polyplexes
Plasmid DNA encoding for firefly luciferase (pcDNA3Luc) was used for all experiments. The molar polymer-to-DNA (N/P) ratio of the polyplexes varied from 1.5 to 100. A mass per charge of 110 and 330 Da was used for p(DMAEA)-ppz and DNA, respectively.
Polyplexes for the in vitro experiments were prepared at a DNA concentration of 10 μg/ml. The buffers used for the preparation of the polyplexes had either a low ionic strength (5 mM Hepes, pH 7.4 or 20 mM Hepes, 5% g/v glucose, pH 7.4, HBG) or a high ionic strength (20 mM Hepes, 150 mM NaCl, pH 7.4, HBS). Polyplexes were prepared by adding four volumes of the polymer solution to one volume of the DNA solution. The resulting dispersions were vortexed and incubated for 30 min at room temperature before use (26).
Polyplexes for the in vivo studies, and for the in vitro erythrocyte aggregation assays were prepared at a DNA concentration of 150 μg/ml (20 mM Hepes, 5% g/v glucose, pH 7.1) by adding one volume of a polymer solution to one volume of DNA solution. The resulting dispersions were vortexed and incubated for 30 min at room temperature. Then, one tenth a volume of a 50% g/v D-glucose stock was added to obtain iso-osmolarity (27).
Characterization of the Polyplexes
The average hydrodynamic diameter and the zeta-potential of the polyplexes were determined using dynamic light scattering and electrophoretic mobility measurements, respectively, at a DNA concentration of 10 μg/ml. The size of the polyplexes was determined using an ALV CGS-3 system (Malvern Instruments, UK). The zeta-potential of the polyplexes was determined using a Zetasizer 2000 (Malvern instruments, UK). Both instruments were calibrated using polystyrene latex beads of defined size and zeta-potential.
Condensation of DNA by the polymers was determined using an ethidium bromide (EtBr) fluorescence quenching assay (28). EtBr was added to a DNA solution 10 min prior to polyplex formation at a molar ratio of 1:8. Next, EtBr labeled polyplexes were prepared at a DNA concentration of 10 μg/ml in Hepes or HBS, as described earlier. The resulting fluorescence was quantified using a Fluorstar Optima plate-reader (BMG Lab Tech, Germany). Fluorescence values were measured at an excitation/emission wavelength of 500 and 600 nm, respectively, and converted into relative, residual fluorescence (Fr) using the following equation:
where Fobs is the observed fluorescence of the polyplex samples, Fe is the fluorescence of EtBr in the absence of DNA, and F0 is the fluorescence of EtBr in the presence of DNA.
In Vitro Transfection
Neuro 2 A (Murine Neuroblastoma) cells (ATCC CCL-131) were cultured in RPMI 1640 medium completed with bovine calf serum (10%) and antibiotics/antimycotics (100 U/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B) at 37°C at a 5% CO2 humidified atmosphere.
Transfection and cytotoxicity studies were carried out in serum containing medium, as described earlier (5). In brief, cells were seeded in a 96-well plate at a density of 1 × 104 cells/well. After twenty-four hours, cells were overlaid with fresh medium (100 μl) and subsequently overlaid with a polyplex dispersion (100 μl, 1 μg DNA, HBS or HBG). Cells were incubated with the polyplexes for 1 h. Subsequently, the medium was refreshed and the cells were cultured for another 24 h. All transfection experiments were carried out in duplicate in two separate 96-well plates. One series was tested for reporter gene expression (luciferase activity) (5), the other series was tested for cell viability using an XTT colorimetric assay (29).
Luciferase activity was measured after lysis of the cell pellet by the addition of 100 μl reporter gene lysis buffer followed by a single freeze/thaw cycle. Luciferase activity of the samples was determined using a Berthold 9507 Luminometer (EG&G Benelux BV, The Netherlands) by mixing 20 μl of the defrosted cell lysate with 100 μl of luciferase assay reagent. Relative light units (RLU) were measured for 10 s at room temperature and were corrected for background values. In this setting, 6 × 103 RLUs corresponded to 1 pg of recombinant luciferase.
The percentage of viable cells was tested using an XTT colorimetric assay, based on the metabolic, cellular reduction of a tetrazolium reagent (29). Cell viability values were normalized to values obtained after incubation with buffer only.
Polyplex-induced Erythrocyte Aggregation
Mouse erythrocytes were isolated from heparinized blood by centrifugation. The erythrocyte pellet was washed several times with HBS buffer. Subsequently, a suspension was prepared by diluting the erythrocyte pellet 40 times with HBS. To 20 μl of the different polyplex dispersions (150 μg/ml DNA in 20 mM Hepes, 5% g/v glucose, pH 7.1, N/P ratio 12), 230 μl of the erythrocyte suspension was added. After 1 h of incubation at 37°C, aliquots of the suspensions were taken for microscopic analysis (20×).
In Vivo Gene Expression Studies
The animal experiments were performed according to national regulations and approved by the local animal experiments ethical committee. Six-week old male A/J mice (Harlan, The Netherlands) were inoculated with 1 × 106 Neuro 2 A cells in the left flank. Eight to twelve days after inoculation, at an average tumor volume of 0.4–0.8 cm3, the different polyplex dispersions were injected into the tail-vein of the mice (0.2 ml, corresponding with 30 μg DNA). The mice were sacrificed 24 h after injection by cervical dislocation and luciferase levels of the different organs were assessed as previously described (30). Briefly, organs were homogenized in 0.5–1 ml of reporter gene lysis buffer, using a tissue homogenizer. The tissue homogenates were incubated on ice for 30 min, then vortexed and subsequently centrifuged at 12,000 g for 10 min. The luciferase activity of the samples was determined using a Berthold 9507 Luminometer by mixing 20 μl of the supernatant with 100 μl of luciferase assay reagent. Relative light units (RLU) were measured for 10 s at room temperature and were corrected for background values. In this setting, 6 × 103 RLUs corresponded to 1 pg of recombinant luciferase. The transfection activity of the polyplexes was expressed as RLU/organ or RLU/tumor.
RESULTS AND DISCUSSION
Polymer Synthesis
A batch of polymer p(DMAEA)-ppz was synthesized and purified as previously described (22). The SEC chromatogram of this starting polymer is shown in Fig. 2a. The polymer had a weight average molecular weight of 680 kDa (Table I). The measured molecular weight distribution was around 2 and is a consequence of the method of polymerization (22–24).
Fractionation
Aqueous size exclusion chromatography (SEC) was used for fractionation and analysis of the p(DMAEA)-ppz polymers. The combined, online use of a series of detectors (low and right angle light scattering, viscosity, refractive index) enabled the determination of the absolute molecular weight of the fractionated polymers, as well as their molecular weight distribution. The refractive index increment (dn/dc) of p(DMAEA)-ppz was determined to be 0.159 ml/g.
Figure 2b shows the SEC chromatogram of one of the fractions of p(DMAEA)-ppz (ppz290). The molecular weight distribution of this fractionated polymer was relatively narrow (Mw/Mn 1.2, Table I). The same held true for the other fractionated polymers (Mw/Mn 1.1–1.3). The weight average molecular weight of the different polymers ranged from 130 to 950 kDa which corresponds to hydrodynamic radii ranging from 8 to 26 nm. The dependency of the calculated molecular weight on retention volume was found to be linear for the different polymers, demonstrating good SEC separation conditions. 31P-NMR analysis of the selected fractions showed one peak at 1.7 ppm (22) indicating that no polymer degradation had occurred during the fractionation process.
Biophysical Characterization
Polyplexes with a mean size between 80 to 90 nm were formed upon complexation of the different polymer fractions with plasmid DNA at a relatively low DNA concentration (10 μg/ml) and at low ionic strength (Hepes buffer), independent of the polymer molecular weight (Table II). All polyplexes had a positive surface charge (zeta-potential 25–29 mV). At high ionic strength (HBS), the different polyplexes gradually aggregated. Most likely, aggregation occurs due to the reduction in zeta-potential of the particles which leads to a decrease in interparticle repulsion and colloidal stability. Aggregate formation was observed over a broad range of N/P ratios in the case of the polyplexes based on the smaller molecular weight polymers (ppz290 and ppz130), whereas the higher molecular weight polymers showed aggregate formation only at N/P ratios of 3 to 6 (starting polymer, ppz950 and ppz570, Fig. 3). The low colloidal stability of the polyplexes based on the low molecular weight polymers is in good agreement with earlier findings on PEI, pLL, and pDMAEMA polyplexes (9,14,19). Probably, the low molecular weight polymers have reduced DNA condensation abilities, due to their reduced number of electrostatic interactions, leading to intensified aggregation at high ionic strength. At a relatively high DNA concentration and a low ionic strength (150 μg/ml DNA in HBG), again small particles were formed, though the complexes were somewhat bigger than formed at lower DNA concentration (Table II).
The DNA binding ability of the different p(DMAEA)-ppz’s was determined by measuring the fluorescence remaining upon addition of the different polymers to EtBr labeled DNA (Fig. 4). At low ionic strength (Hepes buffer), at N/P ratios of 6 and higher, the extent of DNA binding was similar for the different p(DMAEA)-ppz’s (20% of the original fluorescence remaining). Complexation of DNA with the starting polymer resulted in a considerably stronger DNA binding (residual fluorescence 9–13%). At high ionic strength (HBS), 50% of the original fluorescence was remaining after incubation with the fractionated polymers (data not shown), pointing to a reduced DNA binding capability. Also, the starting polymer showed a lower DNA binding ability, albeit stronger than the fractionated polymers (35% of the original fluorescence remaining). The DNA binding observed for the starting polymer, as reflected by the quenching of EtBr fluorescence, is similar to values observed for linear PEI (Mw 22 kDa, PEI22). Mixing of the different p(DMAEA)-ppz fractions in equal mass ratios yielded less EtBr quenching than observed for the starting polymer. Most likely, a particular molecular weight fraction with a high DNA binding ability was present within the starting polymer and is lost during fractionation.
Polyplex Cytotoxicity
Cytotoxicity of the different p(DMAEA)-ppz polyplexes was determined using the XTT assay, by comparing the cellular mitochondrial activity of the polyplex treated cells to that of untreated cells. Figure 5 shows the relative cell viability of Neuro 2 A cells, 24 h after transfection with the polyplexes prepared at different N/P ratios. Cytotoxicity positively correlated with polymer molecular weight, in good agreement with earlier findings on PEI, PLL and other polymers (8,12,19,31,32). Polyplexes based on the starting polymer showed significant cytotoxicity, similar to polyplexes based on the high molecular weight polymer fractions (ppz950 and ppz570). Polyplexes based on PEI22 were found to be equally cytotoxic as ppz290 based polyplexes, at the different N/P ratios. At 48 h after transfection, a similar molecular weight correlation was found (data not shown).
Recently, the mechanisms underlying the enhanced cytotoxicity of 27 kDa PLL polyplexes was investigated, in comparison to 2.9 kDa PLL polyplexes (12,33). The 27 kDa PLL was found to initiate a stronger, early stage, cell membrane perturbation than the low molecular weight polymer, possibly as a result of the more extensive interactions with the cell surface. Moreover, only the 27 kDa PLL and not the 2.9 kDa PLL induced a direct release of cytochrome C from isolated mitochondria, a phenomenon related with late stage cytotoxicity (12). Similar factors might underlie the enhanced cytotoxicity observed for the high molecular weight p(DMAEA)-ppz’s used in this study.
In Vitro Transfection Activity
The in vitro transfection activity of the different p(DMAEA)-ppz polyplexes was investigated. Figure 6 shows the luciferase expression by Neuro 2 A cells, 24 h after transfection with the polyplexes prepared at different N/P ratios. The maximum degree of gene expression was about two orders of magnitude lower than that observed for polyplexes based on PEI22 (N/P 6). Transfection with polyplexes based on the 290 kDa p(DMAEA)-ppz resulted in a bell-shaped curve with highest gene expression levels at N/P ratios of 6 to 50 (Fig. 6d) (19). At low N/P ratios, transfection activity is low, as the polyplexes do not have the favorable physicochemical characteristics required for efficient cellular adhesion, internalization, and a limited extent of (endosomal and nuclear) membrane permeabilization activity (18,19,33). At high N/P ratios, gene expression levels drop most likely due to cytotoxic effects exerted by free, non-DNA associated polymer. In the case of the lowest molecular weight polymer (ppz130), gene expression levels only slightly decrease even at high N/P ratios (Fig. 6e). The low cytotoxicity of the polyplexes based on this polymer enables gene expression even at these high N/P ratios. Transfection with polyplexes based on the starting polymer and the highest molecular weight fraction ppz950 resulted in reduced transfection activity at the intermediate N/P ratios of 6 to 50. Likely, pronounced cytotoxicity at these N/P ratios opposes the transfection process (see polyplex cytotoxicity). Complexes based on ppz570 displayed a somewhat intermediate gene expression profile (Fig. 6c).
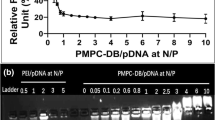
Polyplexes based on the starting polymer and on ppz950 showed a striking maximum in transfection activity at an N/P ratio of 3 (Fig. 6a and b). To study this phenomenon in more detail, polyplexes based on the starting polymer were prepared in transfection medium with high ionic strength (HBS) and in transfection medium with low ionic strength (HBG). In contrast to the polyplexes prepared in HBS, which rapidly aggregated at N/P ratios of 3 to 6 (diameter 800 to >1000 nm, see also Fig. 3), polyplexes prepared in HBG remained small in time (diameter ∼ 100 nm at all N/P ratios > 1.5). Transfection activity of the ‘HBS’ aggregated polyplexes was 15–25 fold higher than their non-aggregated counterparts (Fig. 7). Most likely, complex aggregation in the transfection medium (HBS) is the cause for the observed peak in transfection activity (at N/P 3 to 6, Fig 6a). Earlier, aggregate formation has been found to improve transfection efficiency, likely due to complex sedimentation which enhances the cellular contact and subsequent cellular internalization (34,35). Therefore, likely, the polyplex aggregation observed for the different polymers contributes to the different in vitro gene expression profiles, observed in Fig. 6.
Effect of ionic strength on the in vitro transfection activity of DNA/p(DMAEA)-ppz complexes. Transfection values of polyplexes based on the starting polymer, prepared at high ionic strength (HBS) relative to values determined for polyplexes prepared at low ionic strength (HBG), 24 h after transfection of Neuro 2 A cells (mean, n = 3).
Safety Aspects
Polyplex-induced erythrocyte aggregation is an important factor determining the toxicity of polyplexes after intravenous administration (36). The extent of erythrocyte aggregation was studied, 1 h after incubation with the different polyplexes (150 μg/ml DNA, size 130–230 nm, Table II). At an N/P ratio of 12, the degree of erythrocyte aggregation was found to correlate with the molecular weight of the polymer used. Extensive aggregation was observed for polyplexes based on the starting polymer and the high molecular weight fractions (ppz950 and ppz570) (Fig. 8a–c). Incubation of the erythrocytes with polyplexes based on the lower molecular weight polymers (ppz290 and ppz130) clearly resulted in a more limited erythrocyte aggregation (Fig. 8d and e). For all polymers, the extent of aggregation was found to be positively correlated with the N/P ratio applied, indicating that free polymer contributes to erythrocyte aggregation (36,37).
The extent of polyplex-induced erythrocyte aggregation correlated with the toxicity observed after intravenous administration of the different p(DMAEA)-ppz polyplexes in mice. Tail-vein administration of polyplexes of the starting polymer or the high molecular weight polymers ppz950 and ppz570 was limited to those with N/P ratios of 6 and lower, as lethal toxicity occurred at higher N/P ratios. Remarkably, polyplexes based on the lower molecular weight polymers ppz290 and ppz130 could be administered at two to three fold higher N/P ratios (12 and 20) without causing any apparent toxicity. Polyplex-induced erythrocyte aggregation and subsequent capillary occlusion is the likely mechanism linking polymer molecular weight and in vivo toxicity. An association between polymer molecular weight, erythrocyte aggregation and toxicity after intravenous administration has been suggested earlier for PEI polyplexes (38).
In Vivo Transfection
Tumor and organ gene expression was studied, 24 h after administration of the polyplexes into the tail vein of tumor-bearing mice (N/P ratio of 12 and 20). As a result of occurring toxicity (see safety aspects), gene expression mediated by polyplexes based on the starting polymer and the high molecular weight ppz950 and ppz570 could not be determined at these N/P ratios. At a lower N/P ratio of 6, polyplexes of the latter polymers did not show significant gene expression in any of the organs. Possibly, the absence of gene expression is due to the limited amount of polymer present, leading to complex neutralization or complex dissociation after opsonization by serum proteins (1,13). The administration of polyplexes of the lower weight polymers (ppz290 and ppz130) resulted in considerable tumor gene expression, when prepared at an N/P ratio of 20 (Fig. 9a and b). However, gene expression was not only confined to the tumor tissue, but was also observed in liver and lung. At an N/P ratio of 12, intravenous administration of polyplexes based on the lowest molecular weight polymer (ppz130) resulted in gene expression which was confined to the tumor tissue. This tumor selective gene expression is in good agreement with earlier findings on p(DMAEA)ppz based polyplexes. The extent of luciferase expression is similar to that observed for polyplexes based on PEI22 (N/P 6) (5).
Luciferase expression, 24 h after i.v. administration of pDNA/ppz290 complexes (a) and pDNA/ppz130 complexes (b) into mice bearing a Neuro 2 A s.c. tumor (30 μg DNA/mouse). White bars: N/P ratio 12, hatched bars: N/P ratio 20. Luciferase expression is plotted as relative light units per organ (RLU/organ) (mean + SD, n = 4). Average tumor weight was 600 mg.
CONCLUSION
p(DMAEA)-ppz’s with different molecular weights ranging from 130 to 950 kDa and with narrow molecular weight distributions were obtained by size exclusion chromatography. Polyplexes based on these polymers were prepared and tested for biophysical properties, (cyto)toxicity, erythrocyte aggregation and in vitro and in vivo tumor transfection.
Polymer molecular weight affected many aspects important in tumor transfection. By varying the polymer molecular weight, we were able to optimize the transfection efficiency/toxicity ratio of the p(DMAEA)-ppz polyplexes for in vitro and in vivo tumor transfection. This study shows that in order to obtain well-characterized polyplex transfection systems, there is a need to use polymers with defined molecular weights and narrow distributions.
References
A. G. Schatzlein. Non-viral vectors in cancer gene therapy: principles and progress. Anticancer Drugs 12:275–304 (2001).
M. Ogris, and E. Wagner. Targeting tumors with non-viral gene delivery systems. Drug Discov. Today 7:479–485 (2002).
J. Panyam, and V. Labhasetwar. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv. Drug Deliv. Rev. 55:329–347 (2003).
O. Pillai, and R. Panchagnula. Polymers in drug delivery. Curr. Opin. Chem. Biol. 5:447–451 (2001).
H. K. De Wolf, J. Luten, C. J. Snel, C. Oussoren, W. E. Hennink, and G. Storm. In vivo tumor transfection mediated by polyplexes based on biodegradable poly(DMAEA)-phosphazene. J. Control. Release 109:275–287 (2005).
W. T. Godbey, K. K. Wu, and A. G. Mikos. Size matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J. Biomed. Mater. Res. 45:268–275 (1999).
M. Mannisto, S. Vanderkerken, V. Toncheva, M. Elomaa, M. Ruponen, E. Schacht, and A. Urtti. Structure-activity relationships of poly(L-lysines): effects of pegylation and molecular shape on physicochemical and biological properties in gene delivery. J. Control. Release 83:169–182 (2002).
M. D. Shau, S. J. Tseng, T. F. Yang, J. Y. Cherng, and W. K. Chin. Effect of molecular weight on the transfection efficiency of novel polyurethane as a biodegradable gene vector. J. Biomed. Mater. Res. A 77:736–746 (2006).
K. Kunath, A. Von Harpe, D. Fischer, H. Petersen, U. Bickel, K. Voigt, and T. Kissel. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J. Control. Release 89:113–125 (2003).
S. Werth, B. Urban-Klein, L. Dai, S. Hobel, M. Grzelinski, U. Bakowsky, F. Czubayko, and A. Aigner. A low molecular weight fraction of polyethylenimine (PEI) displays increased transfection efficiency of DNA and siRNA in fresh or lyophilized complexes. J. Control. Release 112:257–270 (2006).
T. Bieber, and H. P. Elsasser. Preparation of a low molecular weight polyethylenimine for efficient cell transfection. Biotechniques 30:74–77, 80–81 (2001).
P. Symonds, J. C. Murray, A. C. Hunter, G. Debska, A. Szewczyk, and S. M. Moghimi. Low and high molecular weight poly(L-lysine)s/poly(L-lysine)-DNA complexes initiate mitochondrial-mediated apoptosis differently. FEBS Lett. 579:6191–6198 (2005).
D. Fischer, B. Osburg, H. Petersen, T. Kissel, and U. Bickel. Effect of poly(ethylene imine) molecular weight and pegylation on organ distribution and pharmacokinetics of polyplexes with oligodeoxynucleotides in mice. Drug Metab. Dispos. 32:983–992 (2004).
C. M. Ward, M. L. Read, and L. W. Seymour. Systemic circulation of poly(L-lysine)/DNA vectors is influenced by polycation molecular weight and type of DNA: differential circulation in mice and rats and the implications for human gene therapy. Blood 97:2221–2229 (2001).
P. Dubruel, B. Christiaens, B. Vanloo, K. Bracke, M. Rosseneu, J. Vandekerckhove, and E. Schacht. Physicochemical and biological evaluation of cationic polymethacrylates as vectors for gene delivery. Eur. J. Pharm. Sci. 18:211–220 (2003).
D. Fischer, H. Dautzenberg, K. Kunath, and T. Kissel. Poly(diallyldimethylammonium chlorides) and their N-methyl-N-vinylacetamide copolymer-based DNA-polyplexes: role of molecular weight and charge density in complex formation, stability, and in vitro activity. Int. J. Pharm. 280:253–269 (2004).
D. G. Anderson, A. Akinc, N. Hossain, and R. Langer. Structure/property studies of polymeric gene delivery using a library of poly(beta-amino esters). Mol. Ther. 11:426–434 (2005).
M. Breunig, U. Lungwitz, R. Liebl, C. Fontanari, J. Klar, A. Kurtz, T. Blunk, and A. Goepferich. Gene delivery with low molecular weight linear polyethylenimines. J. Gene Med. 7:1287–1298 (2005).
P. Van De Wetering, J. Y. Cherng, H. Talsma, and W. E. Hennink. Relation between transfection efficiency and cytotoxicity of poly(2-(dimethylamino)ethyl methacrylate)/plasmid complexes. J. Control. Release 49:59–69 (1997).
M. Koping-Hoggard, K. M. Varum, M. Issa, S. Danielsen, B. E. Christensen, B. T. Stokke, and P. Artursson. Improved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomers. Gene Ther. 11:1441–1452 (2004).
D. V. Schaffer, N. A. Fidelman, N. Dan, and D. A. Lauffenburger. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol. Bioeng. 67:598–606 (2000).
J. Luten, J. H. Van Steenis, R. Van Someren, J. Kemmink, N. M. Schuurmans-Nieuwenbroek, G. A. Koning, D. J. A. Crommelin, C. F. Van Nostrum, and W. E. Hennink. Water-soluble biodegradable cationic polyphosphazenes for gene delivery. J. Control. Release 89:483–497 (2003).
H. R. Allcock, M. B. Mcintosh, E. H. Klingenberg, and M. E. Napierala. Functionalized polyphosphazenes: Polymers with pendent tertiary trialkylamino groups. Macromolecules 31:5255–5263 (1998).
A. N. Mujumdar, S. G. Young, R. L. Merker, and J. H. Magill. A Study of Solution Polymerization of Polyphosphazenes. Macromolecules 23:14–21 (1990).
X. Jiang, A. Van Der Horst, M. J. Van Steenbergen, N. Akeroyd, C. F. Van Nostrum, P. J. Schoenmakers, and W. E. Hennink. Molar-mass characterization of cationic polymers for gene delivery by aqueous size-exclusion chromatography. Pharm. Res. 23:595–603 (2006).
P. Van De Wetering, J. Y. Cherng, H. Talsma, D. J. A. Crommelin, and W. E. Hennink. 2-(Dimethylamino)ethyl methacrylate based (co)polymers as gene transfer agents. J. Control. Release 53:145–153 (1998).
M. Kursa, G. F. Walker, V. Roessler, M. Ogris, W. Roedl, R. Kircheis, and E. Wagner. Novel shielded transferrin-polyethylene glycol-polyethylenimine/DNA complexes for systemic tumor-targeted gene transfer. Bioconjug. Chem. 14:222–231 (2003).
M. X. Tang, and F. C. Szoka. The influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexes. Gene Ther. 4:823–832 (1997).
D. A. Scudiero, R. H. Shoemaker, K. D. Paull, A. Monks, S. Tierney, T. H. Nofziger, M. J. Currens, D. Seniff, and M. R. Boyd. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 48:4827–4833 (1988).
F. Verbaan, I. Van Dam, Y. Takakura, M. Hashida, W. Hennink, G. Storm, and C. Oussoren. Intravenous fate of poly(2-(dimethylamino)ethyl methacrylate)-based polyplexes. Eur. J. Pharm. Sci. 20:419–427 (2003).
M. Neu, D. Fischer, and T. Kissel. Recent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivatives. J. Gene Med. 7:992–1009 (2005).
I. R. Hill, M. C. Garnett, F. Bignotti, and S. S. Davis. In vitro cytotoxicity of poly(amidoamine)s: relevance to DNA delivery. Biochim. Biophys. Acta 1427:161–174 (1999).
S. M. Moghimi, P. Symonds, J. C. Murray, A. C. Hunter, G. Debska, and A. Szewczyk. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol. Ther. 11:990–995 (2005).
D. Fischer, T. Bieber, Y. Li, H. P. Elsasser, and T. Kissel. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm. Res. 16:1273–1279 (1999).
L. Wightman, R. Kircheis, V. Rossler, S. Carotta, R. Ruzicka, M. Kursa, and E. Wagner. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. J. Gene Med. 3:362–372 (2001).
E. Moreau, M. Domurado, P. Chapon, M. Vert, and D. Domurad. Biocompatibility of polycations: in vitro agglutination and lysis of red blood cells and in vivo toxicity. J. Drug Target. 10:161–173 (2002).
S. Boeckle, K. Von Gersdorff, S. Van Der Piepen, C. Culmsee, E. Wagner, and M. Ogris. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J. Gene Med. 6:1102–1111 (2004).
R. Kircheis, L. Wightman, A. Schreiber, B. Robitza, V. Rossler, M. Kursa, and E. Wagner. Polyethylenimine/DNA complexes shielded by transferrin target gene expression to tumors after systemic application. Gene Ther. 8:28–40 (2001).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
de Wolf, H.K., de Raad, M., Snel, C. et al. Biodegradable Poly(2-Dimethylamino Ethylamino)Phosphazene for In Vivo Gene Delivery to Tumor Cells. Effect of Polymer Molecular Weight. Pharm Res 24, 1572–1580 (2007). https://doi.org/10.1007/s11095-007-9299-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-007-9299-z