Abstract
Medulloblastoma is the most common malignant brain tumor of childhood. Despite numerous advances, clinical challenges range from recurrent and progressive disease to long-term toxicities in survivors. The lack of more effective, less toxic therapies results from our limited understanding of medulloblastoma growth. Although TP53 is the most commonly altered gene in cancers, it is rarely mutated in medulloblastoma. Accumulating evidence, however, indicates that TP53 pathways are disrupted in medulloblastoma. Wild-type p53-induced phosphatase 1 (WIP1 or PPM1D) encodes a negative regulator of p53. WIP1 amplification (17q22-q23) and its overexpression have been reported in diverse cancer types. We examined primary medulloblastoma specimens and cell lines, and detected WIP1 copy gain and amplification prevalent among but not exclusively in the tumors with 17q gain and isochromosome 17q (i17q), which are among the most common cytogenetic lesions in medulloblastoma. WIP1 RNA levels were significantly higher in the tumors with 17q gain or i17q. Immunoblots confirmed significant WIP1 protein in primary tumors, generally higher in those with 17q gain or i17q. Under basal growth conditions and in response to the chemotherapeutic agent, etoposide, WIP1 antagonized p53-mediated apoptosis in medulloblastoma cell lines. These results indicate that medulloblastoma express significant levels of WIP1 that modulate genotoxic responsiveness by negatively regulating p53.
Similar content being viewed by others
Introduction
Medulloblastoma is the most common malignant brain tumor of childhood [1, 2]. Treatment with surgery, radiation, and chemotherapy successfully cures many patients, but survivors can suffer significant long-term toxicities affecting their neurocognitive and growth potential [3]. Despite clinical advances, up to 30% of children with medulloblastoma experience tumor progression or recurrence, for which no curative therapy exists. The lack of more effective, less toxic therapies stems from our imperfect understanding of the molecular processes that underlie medulloblastoma growth.
Among the most common cytogenetic lesions affecting medulloblastoma are the gain of the long arm of chromosome 17 (17q) and isochromosome 17q (i17q), consisting of 17p deletion with duplication of 17q, in approximately one-third of cases [4, 5]. Because i17q has been described as the sole cytogenetic lesion in certain medulloblastomas, it may represent a primary event rather than an alteration associated with clonal evolution [4]. Investigators have long sought oncogenes on 17q and tumor suppressor genes on 17p, but with limited success [6].
Although the TP53 tumor suppressor gene (17p13.1) is mutated in approximately half of human malignancies, it is rarely mutated in medulloblastoma [7–9]. However, several lines of evidence suggest that the p53 pathway is perturbed in medulloblastoma. Frank et al. have described activation of the p53-p14ARF pathway in the large cell/anaplastic variant of medulloblastoma [10, 11]. Our collaborators have noted significant nuclear p53 immunopositivity consistent with its activation in approximately 50% of primary human medulloblastoma examined (Adekunle Adesina, personal communication). Deletion of the murine homolog, Trp53, in the Patched haploinsufficient (Ptch+/−) mouse model increases the incidence of spontaneous medulloblastoma from approximately 15 to 100% [12]. Individuals with Li–Fraumeni syndrome who carry germline TP53 mutations are at increased risk for developing medulloblastoma, but fewer than 10% of sporadic medulloblastoma display TP53 mutations [4, 5].
The activity of p53 may be abrogated by alternate mechanisms as observed in other cancers with wild-type TP53 [7]. One mechanism by which p53 function is limited is through amplification or overexpression of MDM2. MDM2 encodes an E3 ubiquitin ligase that normally functions in a negative feedback loop to down-regulate p53 expression, and is amplified in approximately 7% of human malignancies that lack mutation of TP53 [13]. However, Adesina et al. and Batra et al. have documented the absence of MDM2 amplification in medullobastoma [14, 15]. Although MDM2 overexpression has been detected in adult patients [16], there is no functional data to implicate MDM2 in p53 inactivation in medulloblastoma, suggesting alternative mechanisms.
The p53-induced proto-oncogene, WIP1 (wild-type p53-induced phosphatase 1 or protein phosphatase, magnesium-dependent 1, delta, PPM1D) maps to 17q22-q23. Amplification of 17q22-q23 has been described in several cell lines and malignancies including neuroblastoma, breast and ovarian carcinomas, most of which are wild-type for TP53 [17–19]. WIP1 displays p53-dependent oncogenic properties by inactivating p53, p38MAPK, and ATM pathways [17–21]. Previous reports of genomic analysis of medulloblastoma have implicated the gain of the WIP1 locus in its overexpression in a subset of tumors [22, 23].
We present data that confirm prior observations and also define the effects of WIP1 on medulloblastoma cell growth. We have quantitated WIP1 overexpression in primary medulloblastoma specimens and cell lines previously analyzed by comparative genomic hybridization (CGH) [24]. Using fluorescence in situ hybridization (FISH), we determined that WIP1 copy gain and amplification is prevalent among but not exclusively found in the tumors with 17q gain and i17q. We surveyed 33 primary medulloblastoma specimens for WIP1 expression and determined that RNA levels were significantly higher in 16 tumors with 17q gain or i17q. Similarly, our Western immunoblot analysis confirmed significant levels of WIP1 protein expression in primary tumors, generally higher in those with 17q gain or i17q. Genotoxic stress induces p53 phosphorylation and WIP1 expression in D283Med and Daoy medulloblastoma cell lines. Transient transfections indicate that WIP1 antagonizes p53-mediated cell cycle arrest and apoptosis in response to the chemotherapeutic agent, etoposide. These results support the hypothesis that medulloblastoma express significant levels of WIP1 that modulate genotoxic responsiveness by negatively regulating p53.
Materials and methods
Medulloblastoma tissue and cell lines
Medulloblastoma samples (n = 33) were obtained from children diagnosed at Texas Children’s Hospital, Houston, TX, between 1996 and 2004, following informed consents. Tissues were snap-frozen and stored in liquid nitrogen until processed. All specimens were obtained at the time of diagnosis prior to radiation or chemotherapy and subjected to histopathologic review according to WHO criteria [1]. Seven tumors displayed classic histology, 11 were uniformly desmoplastic, nodular, or well-differentiated, seven were predominantly anaplastic or large cell, an additional seven displayed some degree of anaplasia or large cell histology, and one was regarded as none of the above.
Ten patients were female and 23 were male. The mean age at diagnosis was 82 months (median 79 months) with four patients less than 36 months old (range: 12–216 months). The median and mean time of follow-up was approximately 42 months from diagnosis. The majority of patients were M0 (non-metastatic), while one was M2 and eight were M3 stage at diagnosis. All studies were performed according to the institutional guidelines of Texas Children’s Hospital, and with the approval of the Institutional Review Board of Baylor College of Medicine.
The breast carcinoma cell line MCF-7 with known high-level WIP1 amplification, the un-amplified breast epithelial cell line MCF-10A, and the medulloblastoma cell lines, Daoy and D283Med (American Type Culture Collection, Manassas, VA) were maintained as previously described in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA) with high glucose (6 g/l), 2 mM l-glutamine and 10% (vol/vol) heat-inactivated fetal calf serum (Invitrogen, Carlsbad, CA) at 37°C in 5% CO2. Etoposide (VP-16) and cisplatin (CDDP) were applied to cell cultures as media-diluted stock solutions in PBS and DMSO, respectively (Sigma, St. Louis, MO). Ultraviolet (UV) irradiation was performed by exposing cell cultures using a GS Gene Linker UV Chamber (Bio-Rad Laboratories, Hercules, CA).
Fluorescence in situ hybridization (FISH) analysis
Primary medulloblastoma cells were harvested from first passage cultures. Chromosomes were prepared according to standard methods, including confirmatory karyotyping with G-banding [25]. Eleven first or second passage cultures of primary tumors were harvested when cultures were still sub-confluent. The BAC containing the WIP1 locus at chromosome 17q23 (RP11-67D12) and a plasmid containing chromosome 17 centromeric sequences (pZ17-14, generously provided by Dr. Mariano Rocchi) were fluorescently labeled with Spectrum Green and Spectrum Orange, respectively (Vysis, Downers Grove, IL), by nick translation and hybridized to metaphase or interphase spreads of cell lines (MCF-7, Daoy, D283) and primary tumor specimens [26]. We confirmed the map position of WIP1 on normal human metaphase spreads by FISH, as described previously [27]. The slides were counterstained with DAPI, and the images were captured using the Quips Pathvysion System (Applied Imaging, Santa Clara, CA). To determine the WIP1 copy number status, we analyzed 100–200 individual interphase nuclei for each case. The presence of copy gain or amplification was indicated if more than 10% of tumor nuclei displayed an increased copy number relative to the chromosome 17 centromeric probe signals and/or tumor ploidy. In cases in which there was a discrepancy between the number of centromeric signals and ploidy, the ploidy number was used to estimate the relative copy number.
Quantitative real-time RT-PCR (qRT-RTPCR) analysis
Total cellular RNA was prepared from frozen tumor tissue with either Trizol (Invitrogen) or an RNeasy kit (Qiagen, Valencia, CA) depending upon relative abundance, according to the manufacturers’ recommendations. Integrity and concentration of RNA were verified on the Baylor College of Medicine, Houston, TX Microarray Core Facility Bioanalyzer (Agilent Technologies, Santa Clara, CA). Total cellular RNA was reverse transcribed with SuperScript First-Strand Synthesis (Invitrogen) primed with oligo-(dT)12. PCR reactions were performed in a Bio-Rad iCycler using iQ Syber Green Supermix (Bio-Rad Laboratories), and primers listed below. Each PCR reaction was performed for 40 cycles, and performed in triplicate. Primer sequences included: WIP1 sense, 5′-ATCCGCAAAGGCTTTCTCGCTT-3′ and antisense, 5′-TTGGCCATTCCGCCAGTTTCTT-3′; TP53 sense, 5′-CCATCTACAAGCAGTCACAGC-3′ and antisense, 5′-GAGTCTTCCAGTGTGAGATG-3′; GAPDH sense, 5′-AAGGTGAAGGTCGGAGTCAA-3′ and antisense, 5′-AATGAAGGGGTCATTGATGG-3′. Expression of target RNA was internally normalized to expression of GAPDH gene and expressed relative to target gene expression in a human fetal brain RNA pool (Stratagene, La Jolla, CA). Human adult cerebellar (Stratagene) and human fetal cerebellar RNA samples (BioChain, Hayward, CA) were also assayed to provide tissue controls. Amplification products were verified by analyzing melting curves, agarose gel electrophoresis, and with direct sequencing of PCR products.
Western immunoblot analysis
Protein was extracted from frozen tissue by solubilization in boiling lysis buffer (0.5% SDS, 50 mM Tris–Cl, pH 8, 5 mM Na2EDTA, 2% β-mercaptoethanol). Lysates were stored at −80°C and separated by SDS-PAGE, transferred to nitrocellulose membranes (Pall Life Sciences, Ann Arbor, MI), and immunoblotted using standard methods. Primary antibodies included: WIP1 monoclonal antibody (Trevigen, Gaithersburg, MD), Phospho-p53 serine-15 (Ser15)-specific monoclonal antibody (Cell Signaling Technology, Danvers, MA), p53 monoclonal antibody (DO-1, Santa Cruz Biotechnology, Santa Cruz, CA), and β-Actin monoclonal antibody (Sigma). Staining with secondary antibody (Alexa Fluor 680-nm-conjugated goat anti-mouse IgG, Invitrogen) was visualized using an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, Nebraska). Intensity of immunostaining was quantitated using ImageQuant 5.2 (Molecular Dynamics, Piscataway, NJ). Expression of target proteins in each sample was internally normalized to β-actin (determined on re-probed blots), relative to expression in the MCF-10A cell line.
Transfections and luciferase reporter assays
Cell lines were transiently transfected with expression and reporter plasmids using Lipofectin according to manufacturer’s recommendations (Invitrogen). Expression plasmids included: the human cytomegalovirus (CMV) immediate-early (IE) promoter-driven pP53-EGFP plasmid, encoding a p53-enhanced green fluorescent protein (GFP) fusion protein (Clontech, Mountainview, CA). Reporter plasmids included the pP21-luciferase construct to detect p53-activated transcription [28] and the actin promoter-driven Renilla luciferase plasmid, pB-Actin-RL, to provide transfection controls [29]. In each transfection, the expression and reporter plasmids were supplemented by empty vector plasmid (pcDNA3.1, Invitrogen) to achieve an equivalent total DNA concentration.
For flow cytometry, the EGFP-expressing pMaxGFP plasmid (Amaxa, Gaithersburg, MD) provided transfection controls. For functional assessment of in vitro p53 activation, cells transfected with luciferase-encoding reporter plasmids were harvested and processed for dual color luciferase assay according to manufacturer’s recommendations (Promega, Madison, WI), and quantitated using a BenchMark Plus plate spectrophotometer (Bio-Rad).
Flow cytometric and terminal deoxynucleotidyl transferase-mediated biotinylated-dUTP nick end-labeling (TUNEL) analysis
We monitored DNA-indices for cell cycle analysis by multi-parametric flow cytometry using standard methods. Analyses were performed using a Becton Dickinson FACScan flow cytometer (BD Biosciences, San Jose, CA) for the detection of cells stained with propidium iodide (PI) and a 488 nm laser with filter combination for fluorescein isothiocyanate (FITC) and GFP. Single cell suspensions were isolated from culture, fixed in methanol, and stained with PI (100 μg/ml in PBS). Each histogram represents 10,000–100,000 cells for measuring DNA-index and cell cycle. Histogram analysis was performed with the CellQuest program (BD Biosciences). We calculated the sub-G0/G1 peak at a position in the hypodiploid area below a DNA-index of one (2n). Because the nucleus becomes fragmented during apoptosis and numerous individual chromatin fragments may be present in a single cell, the percentage of objects with a fractional DNA-content is represented by the sub-G0/G1 peak. Apoptotic nuclei were identified as a sub-diploid DNA peak, and were distinguished from cell debris on the basis of forward light scatter and PI fluorescence. The significance of differences in apoptotic populations was determined using paired Student t-test.
To detect apoptotic nuclei in sub-G0/G1 peaks, a subset of unstained cells was analyzed for DNA-fragmentation using the Frag-EL DNA fragmentation detection kit, according to manufacturer’s recommendations (Oncogene Research Products, Cambridge, MA). Cells were also counter-stained with PI for DNA quantification. FITC signal was detected on one channel in the logarithmic mode, while UV fluorescence (PI) was recorded in the linear mode on a separate channel. For each measurement, at least 10,000 cells were analyzed. We used CellQuest software for multi-parametric calculations and analyses. Cutoff negative and positive cells resulted from FITC-fluorescence isotype control measurements for each sample.
Results
WIP1 DNA copy number is increased in medulloblastomas with 17q gain
We employed FISH to analyze copy numbers of the WIP1 gene in primary medulloblastoma specimens and cell lines previously analyzed by CGH (Fig. 1). Compared to the centromeric chromosome 17 probe, which consistently displayed 2–4 signals in each metaphase or interphase nucleus, the 17q23 BAC probe (RP11-67D12) containing the WIP1 locus revealed a wider range of copy numbers. By FISH, we identified high-level amplification of the WIP1 gene (>50 copies/cell) in all of the MCF-7 cells analyzed (Fig. 1A). This finding is consistent with previously identified amplification of the 17q23 chromosomal region in MCF-7 cells. The D283Med (D283) medulloblastoma cell line, with i17q, displayed low-level amplification of the WIP1 locus with four to nine copies in each interphase spread examined (Fig. 1B). In contrast, the Daoy medulloblastoma cell line with trisomy 17 displayed an average of three copies of WIP1 (Fig. 1C).
WIP1 copy number is increased in medulloblastomas with gain of chromosome 17q. High power micrographs of interphase nuclei of cell lines and primary human medulloblastoma cells hybridized to fluorescently-labeled WIP1 BAC (green) and centromeric chromosome 17 probes (red): (A) MCF-7 nucleus showing numerous WIP1 amplicons; (B) D283Med (D283) nucleus exhibiting low-level WIP1 amplification; (C) Daoy nucleus showing gain of 17; (D–L) interphase nuclei of medulloblastoma cells from nine patients, illustrating examples of (D–H) diploid and near-diploid nuclei, and (I–L) those with low-level amplification of WIP1
Among the 71 tumors analyzed by CGH, 11 primary medulloblastomas were examined for WIP1 amplification by FISH [24]. Six tumors displayed i17q, two showed trisomy 17, one had loss of 17p12-p13 with gain of 17p11.2-q25, and two had no change in chromosome 17 (Table 1). We detected low-level amplification of the WIP1 locus in 7 of 11 primary medulloblastoma specimens, which also had gain of 17q, including four with i17q by G-banding.
Four tumors were diploid or near-diploid overall (Fig. 1D–G), and seven displayed copy number gain of the WIP1 locus (Fig. 1H–L). Our previously reported CGH analysis of 71 medulloblastoma revealed that 46.8% had gain of 17q, including 30% with i17q and 2.6% with amplifications involving 17q, consistent with previous reports [4]. Comparing CGH results with our FISH analysis reveals that WIP1 DNA copy number correlates with gain of 17q (including i17q) by CGH (P < 0.001, Student’s t-test). All the tumors with copy number gain of WIP1 by FISH also had gain of the WIP1 locus on chromosome 17q22-q23 by CGH. WIP1 amplification in medulloblastoma was low-level compared to the high-level amplification observed in MCF-7 cells. These cytogenetic results indicate that low-level amplification of the WIP1 locus are prevalent in a significant proportion of medulloblastoma.
Medulloblastomas with gain of 17q, including i17q, overexpress WIP1 RNA
Using qRT-RTPCR, we determined the expression of WIP1 RNA in cell lines and a larger series of primary medulloblastoma samples. Human fetal cerebellum specimens displayed 2.1-fold greater WIP1 expression (±0.2, SEM) compared to fetal whole brain. Human adult cerebellum expresses 4.0-fold more WIP1 RNA (±1.0) than fetal total brain. WIP1-amplified MCF-7 cells had 14-fold higher levels of WIP1 RNA (±2.6), relative to human fetal brain (Fig. 2A). Daoy cells expressed 0.23-fold WIP1 RNA (±0.054, SEM), compared to fetal brain (Fig. 2A). In contrast, i17q-positive D283 cells displayed 9.8-fold (±1.8) higher expression of WIP1 RNA than human fetal brain.
Medulloblastomas with gain of 17q express significantly higher WIP1 RNA levels. (A) WIP1 RNA expression, by qRT-RTPCR analysis, is significantly higher in the D283 cell line and in primary medulloblastoma specimens with gain of 17q or i17q (+17q/i17q MB, n = 16) by conventional karyotyping and CGH, compared to Daoy, and primary tumors without i17q or gain of 17q (non-17q gain MB, n = 17). For reference, expression in the WIP1-amplified MCF-7 breast cancer cell line is also shown. Columns, mean expression of at least three experiments; error bars, SEM Y-axis, RNA expression normalized to GAPDH expression and relative to human fetal brain. (B) Relative WIP1 RNA expression in individual medulloblastoma specimens (n = 33) assayed by qRT-RTPCR, and listed with their associated chromosome 17 status. (C) Relative WIP1 RNA expression in medulloblastomas with WIP1 amplification (open diamond) is significantly higher than non-amplified tumors (open circle). Abbreviations: ALL MB, mean (±SEM) of all primary human tumors; i17q, isochromosome 17q; +17, gain of chromosome 17; −17, loss of chromosome 17; NC, no change in chromosome 17
We surveyed 33 primary medulloblastoma specimens for WIP1 expression and determined that RNA levels were significantly higher in 16 tumors with 17q gain or i17q (Fig. 2, Table 2). Of the primary medulloblastomas examined by FISH, WIP1-amplified tumors (n = 7) displayed significantly higher RNA levels than non-amplified tumors (n = 4) (27.7-fold (±1.6) vs. 14.1-fold (±4.5) more WIP1 RNA than fetal control; P = 0.050) (Fig. 2C, Table 2). Among the larger set of analyzed specimens, primary medulloblastoma samples displayed 13.0-fold higher levels of WIP1 RNA (±2.1) than human fetal brain (Fig. 2A–B, Table 2). Analyzed separately, tumors with gain of 17q, including i17q, (n = 16) expressed 20.3-fold more WIP1 RNA (±2.1), compared to those without gain of 17q or i17q (n = 17) with only 5.9-fold more WIP1 RNA (±1.0), relative to human fetal brain (Fig. 2A–B, Table 2). In other words, those tumors with gain of 17q, including i17q, expressed 3.4-fold more WIP1 RNA (±0.86) than those without such cytogenetic lesions. The association between WIP1 RNA expression and the presence or absence of 17q gain (including i17q) was statistically significant (P < 0.00001) (Table 2).
Medulloblastomas with gain of 17q overexpress WIP1 protein
A subset of representative tumors with sufficient tissue was analyzed for WIP1 protein expression. Western immunoblot analysis confirmed significant protein expression in lysates from medulloblastoma cell lines and tumor samples, internally normalized to β-actin expression and relative to expression in the MCF-10A cell line (Fig. 3A–B). The medulloblastoma cell line, Daoy, with trisomy 17, expressed 3.1-fold more WIP1 protein (±0.5, SEM) than MCF-10A cells (Fig. 3A–B). The D283 cell line, with i17q, displayed 15.4-fold higher WIP1 levels (±2.0) than MCF-10A (Fig. 3A–B). Since UV irradiation activates p53 by phosphorylation at serine 15 and induces WIP1 expression [30], we used lysates prepared from irradiated D283 cells to provide control specimens with induced p53 and WIP1 proteins. In UV-irradiated D283 cells, p53 levels stabilized with increased phosphorylation at Ser15 (Fig. 3A). The WIP1-amplified breast cancer line, MCF-7, expressed 4.5-fold more WIP1 (±0.4) than MCF-10A cells (Fig. 3A–B).
Medulloblastomas express significant WIP1 protein. (A) Western blots of cell lines and four primary medulloblastoma specimens reveal significant WIP1 and p53 protein expression. UV irradiation (30 J/m2) of D283 increases WIP1 and p53 expression and induces p53 phosphorylation detected as phospho-p53 (Ser15). Breast carcinoma cell lines, WIP1-amplified MCF-7 and non-amplified MCF-10A are shown for reference. Associated chromosome 17 lesions are also labeled. (B) Western analysis of WIP1 protein expression among primary medulloblastoma specimens (n = 10), normalized to internal reference β-actin and relative to expression in the MCF-10A cell line. Abbreviations: all MB, mean (±SEM) of all primary human tumors (n = 10); i17q, isochromosome 17q; +17, gain of chromosome 17; −17, loss of chromosome 17; CNA, copy number aberration
Of the ten primary medulloblastoma specimens analyzed, five displayed gain of 17q or i17q, one had trisomy 17, another had loss of 17p, and three showed no change in chromosome 17 by CGH. Overall, mean WIP1 protein expression was 6.1-fold (±1.1) greater than expression in MCF-10A cells, internally normalized to β-actin (Fig. 3B). As a group, primary tumors exhibited even more WIP1 than the MCF-7 cell line, which is widely employed as an example of WIP1-amplification. Analyzed separately, primary medulloblastomas with 17q or i17q (n = 6) displayed 7.1-fold (±1.5) more WIP1 protein than MCF-10A, compared to non-17q gain tumors (n = 4) with only 4.6-fold (±1.3) levels (Fig. 3B). We also analyzed primary medulloblastoma lysates for expression of total p53 protein and p53 phosphorylation at serine 15 (Ser15) (Fig. 3). Total p53 and phospho-p53 (Ser15) were readily detectable on Western blots, and suggest functional significance. Overall, Western analysis indicated significant levels of WIP1 protein expression in primary tumors, generally higher in those with 17q gain or i17q, which might be responsible for antagonizing p53 activation.
Genotoxic stress induces p53 phosphorylation and WIP1 expression in medulloblastoma cell lines
To determine the functional significance of WIP1 overexpression, we transiently transfected established medulloblastoma cell lines and assayed changes in cell cycle and apoptosis in response to altered levels of WIP1. The D283 and Daoy cell lines differ in their basal WIP1 expression and their TP53 status, providing cell systems for gain-of-function transfection studies. Genotoxic stress in the form of exposure to UV irradiation or the chemotherapeutic agent VP-16 stabilized p53 levels and induced phosphorylation of p53 in D283 and Daoy cells, respectively (Fig. 4A). We examined the effects of increased constitutive WIP1 by transient transfection with a flag-tagged expression plasmid. Flag-tagged WIP1 could be readily distinguished from endogenous WIP1 on Western blots and WIP1 transfection was accompanied by decreased phospho-p53 (Ser15) in treated D283 and Daoy cells that are wild-type and mutant for TP53, respectively (Fig. 4A).
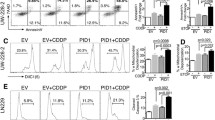
WIP1 antagonizes p53 activation in medulloblastoma cell lines. (A) Western blots of D283 and Daoy medulloblastoma cell lines: Treatment with UV irradiation (UV, 30 J/m2) or etoposide (VP-16, 1 μM) stabilizes wild-type p53 protein and induces phosphorylation of p53 at Ser15. Transfection with the WIP1-flag expression plasmid detected as higher molecular weight band using anti-WIP1 or anti-flag antibodies. Overexpressed WIP1 reduces p53 phosphorylation at serine 15 (Ser15). (B) Increased TP53 and WIP1 RNA in D283 and Daoy cell lines transfected with TP53-GFP and WIP1-flag (WIP1) expression plasmids compared to the MaxGFP control plasmid, as measured by qRT-RTPCR. (C) Luciferase assays reveal that co-transfection of WIP1-flag (WIP1) partially abrogates p53-dependent transcription from the p21-luciferase reporter (relative to constitutively expressed control Renilla-luciferase reporter) in TP53-GFP transfected D283 and Daoy cells (error bars, SEM). (D) TUNEL assay and flow cytometric methods confirm VP-16-induced apoptosis in Daoy and D283 cell lines
Determination of RNA levels with qRT-RTPCR confirmed significant expression of TP53 and WIP1 by transfection with WIP1-flag and TP53-GFP expression plasmids (Fig. 4B). To confirm the transcriptional effects of WIP1- and TP53-expression plasmids, we co-transfected cell lines with the p53-responsive p21-luciferase reporter plasmid and an actin promoter-driven Renilla luciferase plasmid to control for transfection efficiency. Transfected WIP1 partially abrogated p53-dependent luminescence indicating anti-p53 activity in both cell lines in vitro. Presumably because of endogenous wild-type TP53, the relative response in D283 cells was less dramatic than in Daoy cells (Fig. 4C).
VP-16 and constitutively expressed p53 induce apoptosis
Expression from epitope-tagged plasmids enabled us to employ flow cytometric methods to analyze the cell cycle distribution of fluorescently immunolabeled WIP1- and p53-overexpressing transfectants. To stimulate genotoxic responses in vitro, we employed minimally toxic dose concentrations of etoposide (VP-16), a chemotherapeutic topoisomerase II-inhibitor used to treat medulloblastoma clinically. VP-16-induced changes in apoptosis were also determined by flow cytometric quantitation of cells in sub-G0/G1 fractions, which were confirmed by TUNEL analysis (Fig. 4D). Similar results were obtained with micromolar concentrations of another chemotherapeutic agent, cisplatin (data not shown).
When exposed to low concentrations of VP-16, the D283 medulloblastoma cell line underwent increased apoptosis indicated by an increased fraction of cells in sub-G0/G1 by flow cytometric analysis, from 4.5 to 17.2% (P = 0.009) (Fig. 5F). These results indicate that VP-16 induces apoptosis, as described for other cell types with wild-type p53.
WIP1 antagonizes p53-mediated apoptosis and arrest in medulloblastoma cell lines. Flow cytometric analysis of cell cycle distributions of: (A) untreated D283 controls, illustrating the cell populations with 2n DNA content in G0/G1 phase, 4n DNA content in G2/M phase, intermediate DNA content in S phase, and the sub-G0/G1 peak representing apoptotic nuclei; (B) WIP1-flag transfected D283, (C) p53-GFP transfected D283, (D) WIP1-flag and p53-GFP (WIP1 + p53) doubly transfected D283, and (E) etoposide (VP-16)-treated D283 (Y-axis, cell number; X-axis, DNA content). Histograms summarizing flow cytometric analyses of transfected cultures: (F) apoptotic D283 and (G) apoptotic Daoy in Sub-G0/G1, with plasmids and treatment with V-16, as indicated (Y-axis, %; X-axis, transfected plasmid(s) and culture conditions). The following annotation symbols refer to statistically significant differences in sub-G0/G1 cell populations, as determined by paired Student t-test: * D283 cells transfected with p53-GFP plasmid versus mock-transfected D283 cells (P = 0.033). ** D283 cells co-transfected with both p53-GFP and Flag-WIP1 plasmids versus D283 cells transfected with p53-GFP alone (P = 0.026). *** D283 cells transfected with p53-GFP plasmid and treated with VP-16 vs. VP-16-treated, mock-transfected D283 cells (P = 0.019). # D283 cells transfected with Flag-WIP1 plasmid versus untreated, max-GFP control-transfected D283 cells (P = 0.013). ## Daoy cells transfected with p53-GFP plasmid and treated with VP-16 vs. VP-16-treated, mock-transfected Daoy cells (P = 0.044)
Increasing p53 in D283 cells with a GFP-tagged p53 expression plasmid also increased apoptosis (16.9% in sub-G0/G1) compared to mock-transfected controls (4.5%, P = 0.033) (Fig. 5F). VP-16 treatment of p53-transfected D283 cells further increased the sub-G0/G1 population to 25.6% (P = 0.019 compared to mock-transfected VP-16-treated controls) (Fig. 5F). These results suggest that additional constitutive p53 expression in transfected D283 cells can overcome negative regulatory pathways including endogenous MDM2 and WIP1.
We also transfected the TP53-mutant Daoy medulloblastoma cell line. Like wild-type TP53 D283 cells, control Daoy cells treated with VP-16 increased their apoptotic sub-G0/G1 population to 20.4 vs. 6% in untreated controls, suggesting that VP-16 induces apoptosis even in the absence of wild-type p53 (Fig. 5G). Constitutive expression of transfected wild-type p53 in TP53-mutant Daoy cells enhanced VP-16-induced apoptosis to 33.5% (sub-G0/G1), compared to mock-transfected, VP-16-treated controls (14.6%, P = 0.044) (Fig. 5G). These data suggest that transfected p53 expression partially rescues TP53 mutation in Daoy cells.
WIP1 overexpression limits VP-16- and p53-induced apoptosis
We examined the effects of constitutive WIP1 expression in wild-type TP53 D283 cells. VP-16 exposure increased apoptosis to 18.5 from 5.7% in untreated WIP1-transfected D283 (Fig. 5F). Despite limited transfection efficiency, extra WIP1 expression appeared to antagonize p53 activity even in the absence of genotoxic stress. When D283 cells were co-transfected to constitutively overexpress both WIP1 and TP53, they displayed fewer cells in sub-G0/G1 than p53-transfectants (10 vs. 16.9%, respectively; P = 0.026) (Fig. 5F). These data suggest that additional WIP1 limits exogenous p53-mediated apoptosis. In double WIP1- or TP53-transfected D283 cells, VP-16 increased apoptosis (15%) (Fig. 5F). Compared to VP-16-treated p53-transfected D283, double transfectants displayed a trend toward less apoptosis and fewer cells in G0/G1 phase (Fig. 5F). These data are consistent with anti-p53-induced apoptosis, presumably due to WIP1 overexpression, despite genotoxic stimulation.
WIP1-transfected Daoy cells lacking wild-type p53 did not differ in their cell cycle profile from control transfectants under basal conditions, demonstrating the p53-dependence of WIP1 effects. However, VP-16-treated WIP1-transfectants underwent more apoptosis (23.6%) than untreated counterparts (4.1%) (Fig. 5G). These data suggest a minimal effect of constitutively expressed WIP1 in the absence of wild-type p53. Overexpressed WIP1 also limited p53-mediated apoptosis in doubly co-transfected Daoy cells. As in controls, VP-16 treatment of double WIP1/TP53 Daoy transfectants increased cells in sub-G0/G1 (21.8%) (Fig. 5G). However, VP-16-treated double WIP1- or TP53-transfectants displayed fewer apoptotic cells compared to VP-16-treated single p53-transfectants. Thus, excess WIP1 also appears to protect VP-16-treated Daoy cells from p53-induced arrest and apoptosis.
Discussion
Regulation of the responses of medulloblastoma to genotoxic therapies is not well understood. Because TP53 has been implicated but rarely mutated or deleted in medulloblastoma, we focused attention on the candidate oncogene, WIP1. Recently published studies employed genomic methods to associate WIP1 with gain of 17q23 in medulloblastomas. Mendrzyk et al. used array-CGH to identify gain of chromosomal region 17q23.2-qter, a locus containing WIP1, in 24 of 47 (51%) medulloblastomas [22]. WIP1 mRNA was moderately overexpressed with respect to normal cerebellum control in seven of nine tumors. Immunostaining of a large series of tissue micro-arrays revealed strong nuclear expression in 148 of 168 (88%) tumor samples. Ehrbrecht et al. used conventional and array-CGH to identify amplification of 17q23 in three (19%) tumors and copy number gain of 17q23 in another 3 out of 16 medulloblastomas [23]. In 3 of 11 (27%) tumors, WIP1 mRNA was expressed less than fivefold in excess of expression in normal cerebellum control. Using additional approaches, we have confirmed gain of the WIP1 locus in medulloblastomas and quantitated its relative overexpression in our larger series of tumors. These results raise the possibility that overexpressed WIP1 functions as an oncoprotein in medulloblastoma.
WIP1 is a p53-dependent gene induced by genotoxic stress [20, 31]. As a PP2C serine–threonine phosphatase family member, Wip1 can counteract the tumor suppressive activity of p53 through several distinct mechanisms. WIP1 dephosphorylates p53 that has been activated in response to genotoxic stress [21]. WIP1 also dephosphorylates and stabilizes MDM2, promoting p53 degradation (Xiongbin Lu, manuscript submitted). This provides another feedback loop that returns the cell to a homeostatic state following DNA repair. Wip1 can complement different oncogenes in their transformation of mouse embryo fibroblasts. Wip1 overexpression also phenocopies the anti-apoptotic effects of TP53 mutation in vitro and increases tumorgenicity in murine models in vivo [20, 32].
Amplification or overexpression of WIP1 in breast and ovarian cancers may phenocopy the loss of p53 function, thereby contributing to growth and resistance to genotoxic therapies [17, 18]. In breast carcinomas with amplified and overexpressed WIP1, the TP53 locus is rarely mutated, suggesting that WIP1 amplification inhibits p53 activity and reduces selection for p53 mutations during tumor progression [17]. Although WIP1 expression reportedly correlates with clinical outcomes in patients with breast and ovarian carcinomas [33, 34], conclusive multivariate analysis of additional factors (such as the concurrent deletion of a TP53 allele and loss of 17p seen with i17q) awaits future analysis of greater tumor numbers.
Analysis of our series of primary medulloblastomas revealed low-level amplification of WIP1 as well as increased expression of WIP1 mRNA transcript and protein, especially in tumors with 17q gain and i17q. The connection between WIP1 amplification and these common cytogenetic lesions suggests a selective advantage. Since a few tumors with i17q did not display WIP1 amplification, other factors probably contribute to the emergence of this characteristic cytogenetic lesion. Conversely, one or more tumors without 17q gain displayed WIP1 overexpression. Although extra WIP1 copy numbers may explain relative overexpression via gene dosage effects, regulation at the post-transcriptional and -translational levels may also affect WIP1 protein levels. Consistent with our FISH results, qRT-RTPCR and Western blot analyses demonstrated increased levels of WIP1 mRNA and protein; both significantly associated with 17q gain and i17q. The degree of WIP1 RNA overexpression relative to normal cerebellar tissue corresponded to earlier reports [22, 23]. The apparent discrepancy between WIP1 expression at the RNA and protein levels suggests that post-translational modifications also contribute to WIP1 overexpression. The positive and negative regulatory networks appear to involve p38MAPK, MDM2, CHK1, and other DNA damage response factors [21, 30, 33].
Of note, our analysis was limited to those tumor specimens with available fresh tissue for molecular studies and displays an unintended selection bias for mixed tumors with anaplastic features. However, none of our findings appeared to correlate with histologic subtype or clinical features. Having confirmed the gain of WIP1 copy numbers and quantitated its relative overexpression in our medulloblastoma series, we hypothesized that overexpressed WIP1 contributes to treatment resistance by interfering with p53 function in medulloblastoma.
Because the tumor specimens were obtained before genotoxic treatment, their WIP1 and p53 levels do not reflect their activity. Therefore, we employed established cell lines to analyze the effects of overexpression of WIP1. Despite their phenotypic differences, D283 and Daoy cells provide readily manipulable systems for comparing the cell cycle effects of WIP1 in the setting of wild-type versus mutant p53. Our results indicate that chemotherapy-induced genotoxic stress or p53 overexpression alone can induce apoptosis in D283 cells. The constitutive overexpression of p53 overcame endogenous negative regulation in D283 cells, and rescued TP53-mutation in VP-16-treated Daoy cells. These results are consistent with studies of other p53 wild-type cell types, including the neuroepithelial lineage, indicating that VP-16 induces apoptosis in a p53-related manner [35–37].
Medulloblastoma cell lines displayed predicted effects of WIP1 overexpression. In addition to high basal WIP1 levels in D283 cells, additional transfected WIP1 counteracted the apoptotic effects of endogenous p53. The anti-p53 and chemoprotective effects of overexpressed WIP1 were also evident upon VP-16 treatment. These results support the hypothesis that WIP1 overexpression modulates genotoxic responsiveness by negatively regulating p53 in medulloblastoma cells. WIP1 overexpression appears to antagonize p53 activation, and provides medulloblastoma cells the ability to resist arrest and apoptosis due to genotoxic stress.
The frequency of WIP1 amplification and the degree of overexpression in primary medulloblastoma strongly suggests the acquisition of selective advantages that may explain the prevalence of 17q gain and i17q. Based on evidence from medulloblastoma cell lines , we hypothesize that WIP1 regulates p53 function, which contributes to tumor growth and genotoxic treatment resistance. The recent development and characterization of agents that inhibit WIP1 raise the possibility of therapeutic intervention in those medulloblastomas with wild-type TP53 [38, 39].
Abbreviations
- i17q:
-
Isochromosome 17q
- BAC:
-
Bacterial artificial chromosome
- CGH:
-
Comparative genomic hybridization
- FISH:
-
Fluorescence in situ hybridization
- qRT-RTPCR:
-
Quantitative real-time RT-PCR
References
Giangaspero F, Bigner SH, Giordana MT, Kleihues P, Trojanowski JQ (2000) Medulloblastoma. In: Kleihues P, Cavenee WK (eds) Pathology and genetics: tumours of the nervous system. World Health Organization Classification of Tumours. International Agency for Research of Cancer, Lyon, France, pp 96–103
McNeil DE, Cote TR, Clegg L, Rorke LB (2002) Incidence and trends in pediatric malignancies medulloblastoma/primitive neuroectodermal tumor: a SEER update. Surveillance epidemiology and end results Med Pediatr Oncol 39:190–194
Rood BR, Macdonald TJ, Packer RJ (2004) Current treatment of medulloblastoma: recent advances and future challenges. Semin Oncol 31:666–675
Biegel JA (1999) Cytogenetics and molecular genetics of childhood brain tumors. Neuro Oncol 1:139–151
Ellison D (2002) Classifying the medulloblastoma: insights from morphology and molecular genetics. Neuropathol Appl Neurobiol 28:257–282
Cogen PH, McDonald JD (1996) Tumor suppressor genes and medulloblastoma. J Neurooncol 29:103–112
Benard J, Douc-Rasy S, Ahomadegbe JC (2003) TP53 family members and human cancers. Hum Mutat 21:182–191
Ohgaki H, Eibl RH, Wiestler OD, Yasargil MG, Newcomb EW, Kleihues P (1991) p53 mutations in nonastrocytic human brain tumors. Cancer Res 51:6202–6205
Saylors RL III, Sidransky D, Friedman HS, et al (1991) Infrequent p53 gene mutations in medulloblastomas. Cancer Res 51:4721–4723
Frank AJ, Hernan R, Hollander A, et al (2004) The TP53-ARF tumor suppressor pathway is frequently disrupted in large/cell anaplastic medulloblastoma. Brain Res Mol Brain Res 121:137–140
Eberhart CG, Chaudhry A, Daniel RW, Khaki L, Shah KV, Gravitt PE (2005) Increased p53 immunopositivity in anaplastic medulloblastoma and supratentorial PNET is not caused by JC virus. BMC Cancer 5:19
Wetmore C, Eberhart DE, Curran T (2001) Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res 61:513–516
Toledo F, Wahl GM (2006) Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 6:909–923
Adesina AM, Nalbantoglu J, Cavenee WK (1994) p53 gene mutation and mdm2 gene amplification are uncommon in medulloblastoma. Cancer Res 54:5649–5651
Batra SK, McLendon RE, Koo JS, et al (1995) Prognostic implications of chromosome 17p deletions in human medulloblastomas. J Neurooncol 24:39–45
Giordana MT, Duo D, Gasverde S, et al (2002) MDM2 overexpression is associated with short survival in adults with medulloblastoma. Neuro Oncol 4:115–122
Bulavin DV, Demidov ON, Saito S, et al (2002) Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat Genet 31:210–215
Li J, Yang Y, Peng Y, et al (2002) Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet 31:133–134
Saito-Ohara F, Imoto I, Inoue J, et al (2003) PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res 63:1876–1883
Bulavin DV, Phillips C, Nannenga B, et al (2004) Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16(Ink4a)-p19(Arf) pathway. Nat Genet 36:343–350
Lu X, Nannenga B, Donehower LA (2005) PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev 19:1162–1174
Mendrzyk F, Radlwimmer B, Joos S, et al (2005) Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J Clin Oncol 23:8853–8862
Ehrbrecht A, Muller U, Wolter M, et al (2006) Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J Pathol 208:554–563
De Bortoli M, Castellino RC, Lu XY, et al (2006) Medulloblastoma outcome is adversely associated with overexpression of EEF1D, RPL30, and RPS20 on the long arm of chromosome 8. BMC Cancer 6:223
Verma RS, Babu A (eds) (1995) Human chromosomes: principles and techniques. McGraw-Hill, New York, NY
Rauta J, Alarmo EL, Kauraniemi P, Karhu R, Kuukasjarvi T, Kallioniemi A (2006) The serine-threonine protein phosphatase PPM1D is frequently activated through amplification in aggressive primary breast tumours. Breast Cancer Res Treat 95:257–263
Rao PH, Murty VV, Gaidano G, Hauptschein R, Dalla-Favera R, Chaganti RS (1994) Subregional mapping of 8 single copy loci to chromosome 6 by fluorescence in situ hybridization. Cytogenet Cell Genet 66(4):272–273
el-Deiry WS, Tokino T, Velculescu VE, et al (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825
Tischer E, Mitchell R, Hartman T, et al (1991) The human gene for vascular endothelial growth factor. Multiple protein forms are encoded through alternative exon splicing. J Biol Chem 266:11947–11954
Takekawa M, Adachi M, Nakahata A, et al (2000) p53-inducible wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. EMBO J 19:6517–6526
Fiscella M, Zhang H, Fan S, et al (1997) Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci USA 94:6048–6053
Demidov ON, Kek C, Shreeram S, et al (2006) The role of the MKK6/p38 MAPK pathway in Wip1-dependent regulation of ErbB2-driven mammary gland tumorigenesis. Oncogene 26:2502–2506
Zhang X, Kim J, Ruthazer R, et al (2006) The HBP1 transcriptional repressor participates in RAS-induced premature senescence. Mol Cell Biol 26:8252–8266
Hirasawa A, Saito-Ohara F, Inoue J, et al (2003) Association of 17q21-q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin Cancer Res 9:1995–2004
Attardi LD, de VA, Jacks T (2004) Activation of the p53-dependent G1 checkpoint response in mouse embryo fibroblasts depends on the specific DNA damage inducer. Oncogene 23:973–980
Clifford B, Beljin M, Stark GR, Taylor WR (2003) G2 arrest in response to topoisomerase II inhibitors: the role of p53. Cancer Res 63:4074–4081
Nam C, Yamauchi H, Nakayama H, Doi K (2006) Etoposide induces apoptosis and cell cycle arrest of neuroepithelial cells in a p53-related manner. Neurotoxicol Teratol 28:664–672
Belova GI, Demidov ON, Fornace AJ Jr, Bulavin DV (2005) Chemical inhibition of Wip1 phosphatase contributes to suppression of tumorigenesis. Cancer Biol Ther 4:1154–1158
Yamaguchi H, Durell SR, Feng H, Bai Y, Anderson CW, Appella E (2006) Development of a substrate-based cyclic phosphopeptide inhibitor of protein phosphatase 2Cdelta, Wip1. Biochemistry 45:13193–13202
Acknowledgments
We thank Darlene G. Skapura, Linda L. Lin, Diana Joo Youn Hwang, Sowmya Paturi, Jessen A. Rajan, Adekunle Adesina, Lazlo Perlaky, and Texas Children’s Cancer Center Flow Cytometry Core Laboratory for technical assistance; Donald L. Durden for helpful discussions; Ching Lau for supervising tissue procurement; and Mariano Rocchi for providing the chromosome 17 centromeric clone.
This work was supported by funding from: Stephanie Lee Kramer/American Brain Tumor Association Translational Grant (Robert C. Castellino); Alex’s Lemonade Stand Foundation Young Investigator Award (Robert C. Castellino); Associazione Italiana contro le Leucemie (Massimiliano De Bortoli); John S. Dunn Research Foundation; NIH Grants HD042977 (Robert C. Castellino), CA100420 (Lawrence A. Donehower), and NS043517 (John Y. H. Kim); Baylor College of Medicine Collaborative Research (John Y. H. Kim) and Cancer Center Pilot Grants (Lawrence A. Donehower, John Y. H. Kim); The Brain Tumor Society (John Y. H. Kim); John S. McDonnell Foundation (John Y. H. Kim); and John and Carroll Goodman (John Y. H. Kim). AACR-Aflac, Inc. Career Development Award for Pediatric Cancer Research (Robert C. Castellino).
Author information
Authors and Affiliations
Corresponding author
Additional information
Robert C. Castellino and Massimiliano De Bortoli contributed equally to this manuscript.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Castellino, R.C., De Bortoli, M., Lu, X. et al. Medulloblastomas overexpress the p53-inactivating oncogene WIP1/PPM1D . J Neurooncol 86, 245–256 (2008). https://doi.org/10.1007/s11060-007-9470-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-007-9470-8