Abstract
Fungal keratitis is a worldwide-distributed refractory and potentially blinding ocular infection caused by various fungi. It is necessary to investigate the etiological and epidemiological characteristics of this disease and establish a rapid and specific pathogenic identification method. Here, we isolated and identified fungal pathogens of 275 patients with presumed fungal keratitis from Jilin Province, China, and conducted statistical analyses of epidemiological information. The positive rate of fungal culture was 72.0 %. Fusarium sp. was the most common genus among 210 fungal isolates. The predominant species were Fusarium solani, Aspergillus fumigatus, and Candida glabrata, which accounted for over 50 % of the isolated organisms. Corneal trauma and previous use of drugs were the most important predisposing factors. In addition, a multiplex polymerase chain reaction (PCR) was designed with species-specific primers of the three species that could identify them with amplicons of approximately 330 bp from F. solani, 275 bp from A. fumigatus, and 230 bp from C. glabrata. Additionally, PCR with fungal universal primers and multiplex PCR were performed using DNA prepared by an improved DNA extraction method from corneal scrapings. With this method, fungal pathogens from corneal scrapings could be specifically and rapidly identified within 8 h. The culture-independent rapid identification of corneal scrapings may have great significance for the early diagnosis and treatment of fungal keratitis.
Similar content being viewed by others
Introduction
Fungal keratitis is a refractory and potentially blinding ocular infection with corneal ulceration, suppurative infection, and visual loss [1, 2]. In recent years, the incidence of the disease has increased significantly. It is associated with factors such as corneal trauma, previous use of antibiotics and corticosteroids, wearing contact lenses, and corneal surgery or transplantation. Because this disease is difficult to treat, and relapse is common, it seriously affects the health and life of patients [2–6].
Fungal keratitis is a harmful and rapidly progressive disease caused by various fungi with similar clinical symptoms [1, 2]. Early diagnosis is very difficult to attain, and this condition is easily misdiagnosed because the epidemiological characteristics are unclear. Moreover, corneal scrapings are difficult to collect and have a low detection rate. In addition, the slow growth of the pathogen and ineffective methods of morphological identification are time-consuming. Treatment remains limited by the scarcity of effective antifungal drugs and the serious resistance of some pathogens [1–4]. Therefore, establishing a rapid, specific method to diagnose fungal keratitis, as well as understanding the etiological and epidemiological characteristics of it, is significant for early diagnosis, effective treatment, and prognosis after recovery.
To rapidly diagnose fungal keratitis via culture-independent methods, the etiological and epidemiological characteristics of fungal keratitis in Jilin Province, China, should be determined. An effective DNA extraction method from corneal scrapings and multiplex polymerase chain reaction (PCR) should be established to identify the predominant pathogens.
Materials and Methods
Microbiological Examination and Epidemiological Analysis
The present retrospective study of 275 cases of presumed fungal keratitis was conducted from January 2005 to December 2012 at three affiliated hospitals of Jilin University in Jilin Province, China. Epidemiological data, such as sex, age, seasonal incidence, risk factors, and medication history, were investigated. In addition, a Chi-square test and multivariate logistic regression analyses were performed to determine the relationship between fungal infection and epidemiological variables using SPSS software 16.0. Corneal scrapings were collected and placed in aseptic tubes by an ophthalmologist using standard clinical techniques. All procedures conformed to the provisions of the Declaration of Helsinki. The scrapings were directly cultured in potato dextrose broth (PDB; Becton–Dickinson, Sparks, MD, USA) at 25 °C for 15 days with shaking. The morphological, physiological, and biochemical characteristics of the isolates were identified using conventional microbiology techniques as described previously [7].
Establishment of a Multiplex PCR Method
According to epidemiological findings in this study, Fusarium solani, Aspergillus fumigatus, and Candida glabrata are the predominant pathogens of fungal keratitis in Jilin Province. Three pairs of species-specific primers were designed based on the sequence homology analysis of the mitochondrial cytochrome b (cyt b) gene. They were fFuso1 (5′-CTC TGT TAA TAA TGC AAC TC-3′) and rFuso1 (5′-TGG TAC TAT AGC TGG AGG AG-3′) for F. solani, which produced an amplicon of approximately 330 bp; fAfu2 (5′-CGC TTT AGC TGC ATT AGT AA-3′) and rAfu3 (5′-AAG TAT CAT TCC GGA ACA-3′) for A. fumigatus, which produced an amplicon of 275 bp; and fCgl2 (5′-GCC AAG ATA TTG TAC AAT GAT-3′) and rCgl2 (5′-GAA ATA ACC ATG CAT TCC-3′) for C. glabrata, which produced an amplicon of 230 bp.
Fifty reference fungal strains, including 11 species of Fusarium (15 strains), 11 species of Aspergillus (15 strains), eight species of Candida (14 strains), and six species of other genera (six strains), as well as three representative bacterial strains, were used to establish a multiplex PCR method (Table 1). Human cornea from remaining normal tissue after keratoplasty was used as a control.
Genomic DNA was extracted from fungal mycelia using solutions I, II, and III of the GenTLE™ for yeast kit (Takara Co., Ltd., Shuzo, Japan) as described previously [7]. Genomic DNA from bacterial stains was extracted as follows: A small number of colonies were placed in 50 μL sterile water, heated at 95 °C for 10 min, and centrifuged at 4 °C and 11,300×g for 10 min. Bacterial DNA was thus concentrated in the supernatant. The quality and quantity of DNA were assessed by spectrophotometry (at 260 and 280 nm on Nanodrop) and were determined to be excellent for PCR.
First, monoplex PCRs with each pair of primers were performed to confirm the primer specificity using the Takara PCR amplification kit (Takara Co., Ltd., Dalian, China). Then, multiplex PCR was performed using three pairs of primers. Each PCR was performed in a 50-μL final volume, which contained 20 ng genomic DNA, 5 μL 10 × PCR buffer, 4 μL dNTP mixture (2.5 mM each dATP, dCTP, dGTP, and dTTP), 1.25 U Taq polymerase, and 1 μL of each primer (10 μM each). After initial denaturation at 95 °C for 2 min, amplification was performed for a total of 30 cycles with denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 1 min, and the final extension was at 72 °C for 10 min. Amplicon sizes were confirmed via electrophoresis, and a DL2000 DNA Marker (Takara) was used as a molecular weight marker [7].
Rapid Identification of Corneal Scrapings
The corneal scraping was placed in a 1.5-mL centrifuge tube and ground in liquid nitrogen. Twenty microliters proteinase K (20 mg/mL) was added, and the mixture was digested at 60 °C for 20 min. Then, DNA was extracted using the modified method of the GenTLE™ for yeast kit. The mixture was added to 540 μL solution I and heated at 37 °C for 20 min after thorough mixing. Twenty microliters proteinase K (20 mg/mL) was mixed with the solution, and digestion was performed at 60 °C for 10 min. After adding 60 μL solution II, the mixture was shaken for a few seconds and heated at 95 °C for 15 min. Finally, 300 μL solution III was added, mixed by inversion (not vortex), quickly submerged in ice for 5 min, and centrifuged at 11,300×g for 15 min. Approximately 600 μL supernatant was mixed with an equal volume of isopropanol, placed on ice for 1 h, and centrifuged at 4 °C and 11,300×g for 5 min. The pellet was washed with 300 μL 70 % ice-cold ethanol and centrifuged at 4 °C and 11,300×g for 5 min. Then, the pellet was dried in a centrifugal concentrator for 30 min. Prior to amplification, each DNA sample was diluted with 30 µL sterile, double-distilled water.
Double corneal scrapings from 42 cases of presumed fungal keratitis were collected. One sample from each case was subjected to DNA extraction using the above method, and the other scraping was used for fungal culture. Fungal infection was determined based on PCR amplification results using universal primers E1M4 and rE2M4, as described previously [7, 8]. In addition, the fungal species was determined by multiplex PCR. Statistical analysis using McNemar’s test (SPPS version 16.0) was performed to compare the results from the PCR analysis and the fungal culture of corneal scrapings.
Results
Etiological and Epidemiological Analyses
From 2005 to 2012, 275 patients with suspected fungal keratitis were examined. They came from various regions of Jilin Province. Most of them had typical clinical features, such as corneal superficial feathery infiltration or ulcers, increased secretions, ciliary congestion, hypopyon, and visual impairment. The fungal culture of 198 cases (72.0 %) was positive, of which, 180 (90.9 %) were pure fungal isolates, 12 (6.1 %) were mixed with other fungi, and 6 (3.0 %) were mixed with bacteria.
According to the clinical features and fungal culture, 198 patients were diagnosed with fungal keratitis. Among these patients, 210 fungal isolates belonging to 17 genera and 29 species were isolated and identified. The isolates of Fusarium were the most common (49.5 %), followed by Aspergillus (18.6 %), Candida (12.4 %), and other genera (19.5 %) such as Alternaria, Acremonium, Cladosporium, and Beauveria. Among these isolates, the predominant species were F. solani, A. fumigatus, and C. glabrata, accounting for 56.6 % of the isolated fungi (Table 2).
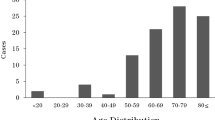
Among the patients, 121 (61.1 %) were male and 77 (38.9 %) were female, and 169 were farmers (85.4 %). The age of patients ranged from 18 to 78 years, with a mean age of 51.6 ± 11.7 years. The patients aged 51–60 years (31.8 %) were particularly susceptible to infection (Fig. 1). Infection often occurred in autumn (September to November, 48.5 %) (Fig. 2). Trauma (28.8 %) and previous use of antibiotics, corticosteroids, or antifungal drugs (58.1 %) were very important predisposing factors. A statistically significant difference (P = 0.005 < 0.01) was observed for the season of onset based on a Chi-square test. The results of multivariate logistic regression analysis (P = 0.013 < 0.05) indicated that the incidence of the disease was associated with the predisposing factors.
Establishment of a Multiplex PCR Method
To confirm the species specificity of the designed primers, monoplex PCRs were performed. The results showed that only F. solani, A. fumigatus, and C. glabrata were amplified with a single, clear, stable, and specific amplicon of approximately 330, 275, and 230 bp, respectively, whereas DNA from other fungi was not amplified (data not shown). These results indicated that the designed primers exhibited species specificity. As a result, multiplex PCR was performed using the three pairs of primers. Only DNA from the three species was amplified with specific amplicons, whereas DNA from other fungi, pathogenic bacteria, and normal human cornea was not amplified (Fig. 3). The amplification accuracy was determined by sequencing and alignment of the amplicons. Triplicate experiments showed that this method was stable and reproducible. These findings suggested that multiplex PCR could specifically and simultaneously identify the three predominant pathogenic fungi.
Results of multiple PCR using three pairs of species-specific primers. Lane M DL2000 DNA Marker, lane 1 F. solani IFM 48449, lane 2 A. fumigatus IFM 40808, lane 3 C. glabrata ATCC90030, lane 4 C. albicans ATCC90028, lane 5 A. flavus IFM 55648, lane 6 F. verticillioides IFM 49276, lane 7 Penicillium chrysogenum JLCC 30780, lane 8 Mucor circinelloides IFM 40507, lane 9 Absidia corymbifera JLCC 30807, lane 10 Alternaria alternata JLCC 33753, lane 11 Curvularia lunata JLCC 31419, lane 12 Staphylococcus aureus ATCC 25923, lane 13 Escherichia coli ATCC 25922, lane 14 Pseudomonas aeruginosa ATCC 27853, lane 15 normal human cornea
Rapid Fungal Identification from Corneal Scrapings
For rapid fungal identification from corneal scrapings, genomic DNA was directly extracted from the specimens and amplified with the universal fungal primers. DNA from 37 cases was amplified with an amplicon of approximately 430 bp, which was the same as that from the control genomic DNA of the cultured strain (Fig. 4). The positive identification rate was 88.1 %. Then, the established multiplex PCR was performed with the DNA from the positive cases. The amplicon was approximately 330 bp from F. solani, 275 bp from A. fumigatus, and 230 bp from C. glabrata. If two or three amplicons were found, then mixed infection occurred. If no amplicon was observed, then other species of fungi were present (Fig. 5). The multiplex PCR results indicated that 14 cases were caused by F. solani, six cases by A. fumigatus, one case by C. glabrata, one case by F. solani mixed with C. glabrata, and the remaining 15 cases by other fungi. Moreover, this method can rapidly identify the predominant pathogens from corneal scrapings within 8 h.
Identification of the positive cases via multiplex PCR. Lane M DL2000 DNA Marker, lane 1 F. solani IFM 48449, lane 2 A. fumigatus IFM 40808, lane 3 C. glabrata ATCC90030, lane 4–15 corneal scrapings. Note Patients of lanes 4, 7, and 9 were infected with F. solani, lanes 6 and 14 were infected with A. fumigatus, lane 12 was infected with C. glabrata, lane 10 was infected with F. solani mixed with C. glabrata, and other lanes were infected with other species of pathogenic fungi
The positive rate of fungal culture of the 42 corneal scraping cases was 69.0 %. This method usually requires 5–7 days (or even longer) to identify isolates. A statistically significant difference was noted between the positive identification rate of these two methods (P = 0.021 < 0.05). These findings suggested that multiplex PCR was superior to fungal culture.
Discussion
Fungal keratitis is a worldwide disease. The prevalence of this disease is related to climate, as well as environmental and geographical conditions, and the incidence of keratitis and its pathogenic spectrum in various countries and regions varies [5, 9]. Some researchers hypothesized that fungal keratitis occurs at a different rate depending on the gross national income of a country [10]. Early diagnosis and prompt effective treatment are limited because of few specimens, a low isolation rate, few antifungal drugs, and an increasing prevalence of drug-resistant strains [1–4, 11]. Therefore, studying etiological and epidemiological characteristics of fungal keratitis, as well as establishing a rapid and specific identification method of predominant pathogens, will be conducive to better prevention, diagnosis, and treatment of this disease.
In developing countries in tropical and subtropical regions such as India, Iran, and Brazil, fungal keratitis accounts for 30–50 % of cases of infectious keratitis, and the predominant pathogens are Fusarium, Aspergillus, Curvularia, and Candida [5, 6, 12]. In the USA, the UK, and other developed countries in temperate regions, the main pathogens are Candida and Fusarium [13, 14]. In China, more than 60 % of infectious keratitis cases are caused by fungi. The genus Fusarium is the most commonly isolated genus, followed by Aspergillus, Penicillium, and Curvularia [15–17].
This study revealed that fungal keratitis in Jilin Province was caused by a wide variety of fungi. The predominant species were F. solani, A. fumigatus, and C. glabrata, which were isolated from over 50 % of the selected cases. C. glabrata isolates were higher in number than C. albicans, which causes Candida keratitis. However, in other studies, C. albicans was the most common pathogen that caused Candida keratitis in the UK and India [18, 19]. The fact that C. glabrata was isolated most frequently in our study may be related to previous drug treatment of most patients before diagnosis. In this study, the isolation rate of C. albicans may be reduced because it is sensitive to azole drugs.
Our epidemiological analysis showed that the incidence was higher in harvest season (September to November) than during other times of the year, corneal trauma and previous drug treatment were the important predisposing factors, and most patients were farmers. These findings may be influenced by the facts that Jilin Province is an important agricultural area in China and that the occurrence of mechanical corneal injury was increased during the harvest season. This process often leads to misdiagnosis and treatment delay because clinicians of the local hospitals lack knowledge of the disease [5, 20]. Therefore, early and accurate diagnosis of the disease is the key to timely and effective treatment and retention of the affected eye.
Currently, the clinical diagnosis of fungal keratitis is dependent on fungal culture. However, the difficulty of collecting corneal scrapings, few specimens, and the long time required for culture (often 1 to 2 weeks, or in some cases, a month or more) often delay early diagnosis and treatment [2, 3]. In recent years, techniques such as PCR, restriction fragment length polymorphism analysis, and reverse line blot hybridization have been used for the identification, diagnosis, and epidemiological studies of fungal keratitis [1–3, 21].
Multiplex PCR, a novel PCR technique, can simultaneously identify various pathogens in one reaction, which helps reduce misdiagnosis. In addition, because multiplex PCR has many advantages, such as suitable specificity, high isolation rate, rapid response, and easy operation, this technique has been widely used for the diagnosis of microbial and parasitic infections, but it has rarely been reported for fungal keratitis [22, 23].
In our study, the etiological characteristics of fungal keratitis in Jilin Province were investigated. F. solani, A. fumigatus, and C. glabrata were confirmed as the predominant species. According to this feature, multiplex PCR was established and species-specific primers were designed and used for pathogenic fungi identification from corneal scrapings. Furthermore, an improved DNA extraction method was used to obtain DNA from corneal scrapings. This is a very useful method that could rapidly identify the three predominant pathogens that caused 56.6 % of infections in Jilin Province at one time. Compared with fungal culture, which required 1 week or more, this method could rapidly and accurately identify fungi from corneal scrapings within 8 h. Additionally, the positive identification rate was significantly higher than that of fungal culture (P < 0.05). These results indicate that the method established in this study can be used as a simple, rapid, and specific method for fungal keratitis diagnosis in conventional laboratories.
In this study, the etiological and epidemiological characteristics of fungal keratitis in Jilin Province, China, were retrospectively studied. In addition, a multiplex PCR method was established for the identification of predominant fungal pathogens. A culture-independent method for the rapid identification of fungal keratitis was developed, providing a scientific basis for early diagnosis and clinical application.
References
Vengayil S, Panda A, Satpathy G, et al. Polymerase chain reaction-guided diagnosis of mycotic keratitis: a prospective evaluation of its efficacy and limitations. Invest Ophthalmol Vis Sci. 2009;50:152–6.
Thomas PA, Kaliamurthy J. Mycotic keratitis: epidemiology, diagnosis, and management. Clin Microbiol Infect. 2013;19:210–20.
Shukla PK, Kumar M, Keshava GB. Mycotic keratitis: an overview of diagnosis and therapy. Mycoses. 2008;51:183–99.
Thomas PA. Current perspectives on ophthalmic mycoses. Clin Microbiol Rev. 2003;16:730–97.
Thomas PA. Fungal infections of the cornea. Eye (Lond). 2003;17:852–62.
Sirikul T, Prabriputaloong T, Smathivat A, Chuck RS, Vongthongsri A. Predisposing factors and etiologic diagnosis of ulcerative keratitis. Cornea. 2008;27:283–7.
He D, Hao J, Zhang B, et al. Pathogenic spectrum of fungal keratitis and specific identification of Fusarium solani. Invest Ophthalmol Vis Sci. 2011;52:2804–8.
Wang L, Yokoyama K, Miyaji M, Nishimura K. Mitochondrial cytochrome b gene analysis of Aspergillus fumigatus and related species. J Clin Microbiol. 2000;38:1352–8.
Kalkanci A, Ozdek S. Ocular fungal infections. Curr Eye Res. 2011;36:179–89.
Shah A, Sachdev A, Coggon D, Hossain P. Geographic variations in microbial keratitis: an analysis of the peer-reviewed literature. Br J Ophthalmol. 2011;95:762–7.
Tu EY, McCartney DL, Beatty RF, Springer KL, Levy J, Edward D. Successful treatment of resistant ocular fusariosis with posaconazole (SCH-56592). Am J Ophthalmol. 2007;143:222–7.
Chowdhary A, Singh K. Spectrum of fungal keratitis in North India. Cornea. 2005;24:8–15.
Galarreta DJ, Tuft SJ, Ramsay A, Dart JK. Fungal keratitis in London: microbiological and clinical evaluation. Cornea. 2007;26:1082–6.
Gower EW, Keay LJ, Oechsler RA, et al. Trends in fungal keratitis in the United States, 2001 to 2007. Ophthalmology. 2010;117:2263–7.
Zhong WX, Sun SY, Zhao J, Shi WY, Xie LX. Retrospective study of suppurative keratitis in 1054 patients. Zhonghua Yan Ke Za Zhi. 2007;43:245–50.
Xie L, Zhong W, Shi W, Sun S. Spectrum of fungal keratitis in north China. Ophthalmology. 2006;113:1943–8.
Shi W, Wang T, Xie L, et al. Risk factors, clinical features, and outcomes of recurrent fungal keratitis after corneal transplantation. Ophthalmology. 2010;117:890–6.
Tuft SJ, Tullo AB. Fungal keratitis in the United Kingdom 2003–2005. Eye (Lond). 2009;23:1308–13.
Sengupta J, Khetan A, Saha S, Banerjee D, Gangopadhyay N, Pal D. Candida keratitis: emerging problem in India. Cornea. 2012;31:371–5.
Chakrabarti A, Singh R. The emerging epidemiology of mould infections in developing countries. Curr Opin Infect Dis. 2011;24:521–6.
Ghosh A, Basu S, Datta H, Chattopadhyay D. Evaluation of polymerase chain reaction-based ribosomal DNA sequencing technique for the diagnosis of mycotic keratitis. Am J Ophthalmol. 2007;144:396–403.
Bu R, Sathiapalan RK, Ibrahim MM, et al. Monochrome Light Cycler PCR assay for detection and quantification of five common species of Candida and Aspergillus. J Med Microbiol. 2005;54:243–8.
Esposto MC, Cogliati M, Tortorano AM, Viviani MA. Determination of Cryptococcus neoformans var. neoformans mating type by multiplex PCR. Clin Microbiol Infect. 2004;10:1092–4.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (No. 30910103903), the Important National Science & Technology Specific Projects of China (No. 2013ZX10004612-006), and the grant from the Science and Technology Department of Jilin Province, China (No. 20130522014JH). We thank Professor Koji Yokoyama (Medical Mycology Research Center of Chiba University, Japan) for his help and guidance. And there is no conflict of interests to declare.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
He, D., Hao, J., Gao, S. et al. Etiological Analysis of Fungal Keratitis and Rapid Identification of Predominant Fungal Pathogens. Mycopathologia 181, 75–82 (2016). https://doi.org/10.1007/s11046-015-9950-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-015-9950-x